IQGAP1 suppresses TβRII-mediated myofibroblastic activation and metastatic growth in liver
- PMID: 23454766
- PMCID: PMC3582119
- DOI: 10.1172/JCI63836
IQGAP1 suppresses TβRII-mediated myofibroblastic activation and metastatic growth in liver
Abstract
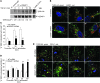
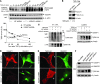
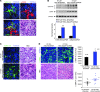
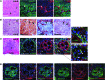
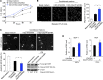
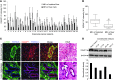
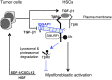
In the tumor microenvironment, TGF-β induces transdifferentiation of quiescent pericytes and related stromal cells into myofibroblasts that promote tumor growth and metastasis. The mechanisms governing myofibroblastic activation remain poorly understood, and its role in the tumor microenvironment has not been explored. Here, we demonstrate that IQ motif containing GTPase activating protein 1 (IQGAP1) binds to TGF-β receptor II (TβRII) and suppresses TβRII-mediated signaling in pericytes to prevent myofibroblastic differentiation in the tumor microenvironment. We found that TGF-β1 recruited IQGAP1 to TβRII in hepatic stellate cells (HSCs), the resident liver pericytes. Iqgap1 knockdown inhibited the targeting of the E3 ubiquitin ligase SMAD ubiquitination regulatory factor 1 (SMURF1) to the plasma membrane and TβRII ubiquitination and degradation. Thus, Iqgap1 knockdown stabilized TβRII and potentiated TGF-β1 transdifferentiation of pericytes into myofibroblasts in vitro. Iqgap1 deficiency in HSCs promoted myofibroblast activation, tumor implantation, and metastatic growth in mice via upregulation of paracrine signaling molecules. Additionally, we found that IQGAP1 expression was downregulated in myofibroblasts associated with human colorectal liver metastases. Taken together, our studies demonstrate that IQGAP1 in the tumor microenvironment suppresses TβRII and TGF-β dependent myofibroblastic differentiation to constrain tumor growth.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

