Ubiquitylation-dependent localization of PLK1 in mitosis
- PMID: 23455478
- PMCID: PMC7116173
- DOI: 10.1038/ncb2695
Ubiquitylation-dependent localization of PLK1 in mitosis
Abstract
Polo-like kinase 1 (PLK1) critically regulates mitosis through its dynamic localization to kinetochores, centrosomes and the midzone. The polo-box domain (PBD) and activity of PLK1 mediate its recruitment to mitotic structures, but the mechanisms regulating PLK1 dynamics remain poorly understood. Here, we identify PLK1 as a target of the cullin 3 (CUL3)-based E3 ubiquitin ligase, containing the BTB adaptor KLHL22, which regulates chromosome alignment and PLK1 kinetochore localization but not PLK1 stability. In the absence of KLHL22, PLK1 accumulates on kinetochores, resulting in activation of the spindle assembly checkpoint (SAC). CUL3-KLHL22 ubiquitylates Lys 492, located within the PBD, leading to PLK1 dissociation from kinetochore phosphoreceptors. Expression of a non-ubiquitylatable PLK1-K492R mutant phenocopies inactivation of CUL3-KLHL22. KLHL22 associates with the mitotic spindle and its interaction with PLK1 increases on chromosome bi-orientation. Our data suggest that CUL3-KLHL22-mediated ubiquitylation signals degradation-independent removal of PLK1 from kinetochores and SAC satisfaction, which are required for faithful mitosis.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

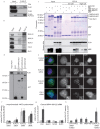
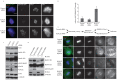

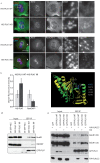

Comment in
-
Cullin' PLK1 from kinetochores.Nat Cell Biol. 2013 Apr;15(4):347-8. doi: 10.1038/ncb2722. Nat Cell Biol. 2013. PMID: 23548928
-
Cell division: a timely exit for PLK1.Nat Rev Mol Cell Biol. 2013 Apr;14(4):194-5. doi: 10.1038/nrm3570. Epub 2013 Mar 13. Nat Rev Mol Cell Biol. 2013. PMID: 23563050 No abstract available.
-
Regulating PLK1 dynamics by Cullin3/KLHL22-mediated ubiquitylation.Cell Cycle. 2013 Aug 15;12(16):2528-9. doi: 10.4161/cc.25839. Epub 2013 Jul 29. Cell Cycle. 2013. PMID: 23907128 Free PMC article. No abstract available.
References
-
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396(6712):643–649. - PubMed
-
- Strebhardt K. Multifaceted polo-like kinases: drug targets and antitargets for cancer therapy. Nat Rev Drug Discov. 2010;9(8):643–660. - PubMed
-
- Petronczki M, Lenart P, Peters JM. Polo on the Rise-from Mitotic Entry to Cytokinesis with Plk1. Dev Cell. 2008;14(5):646–659. - PubMed
-
- Sumara I, et al. Roles of polo-like kinase 1 in the assembly of functional mitotic spindles. Curr Biol. 2004;14(19):1712–1722. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

