Differential innate immune response programs in neuronal subtypes determine susceptibility to infection in the brain by positive-stranded RNA viruses
- PMID: 23455712
- PMCID: PMC3618596
- DOI: 10.1038/nm.3108
Differential innate immune response programs in neuronal subtypes determine susceptibility to infection in the brain by positive-stranded RNA viruses
Abstract
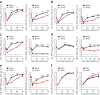
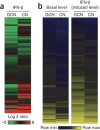
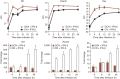
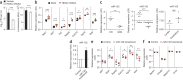
Although susceptibility of neurons in the brain to microbial infection is a major determinant of clinical outcome, little is known about the molecular factors governing this vulnerability. Here we show that two types of neurons from distinct brain regions showed differential permissivity to replication of several positive-stranded RNA viruses. Granule cell neurons of the cerebellum and cortical neurons from the cerebral cortex have unique innate immune programs that confer differential susceptibility to viral infection ex vivo and in vivo. By transducing cortical neurons with genes that were expressed more highly in granule cell neurons, we identified three interferon-stimulated genes (ISGs; Ifi27, Irg1 and Rsad2 (also known as Viperin)) that mediated the antiviral effects against different neurotropic viruses. Moreover, we found that the epigenetic state and microRNA (miRNA)-mediated regulation of ISGs correlates with enhanced antiviral response in granule cell neurons. Thus, neurons from evolutionarily distinct brain regions have unique innate immune signatures, which probably contribute to their relative permissiveness to infection.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
-
- Schoneboom BA, Lee JS, Grieder FB. Early expression of IFN-α/β and iNOS in the brains of Venezuelan equine encephalitis virus–infected mice. J. Interferon Cytokine Res. 2000;20:205–215. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

