Prokaryotic nucleotide excision repair
- PMID: 23457260
- PMCID: PMC3578354
- DOI: 10.1101/cshperspect.a012591
Prokaryotic nucleotide excision repair
Abstract
Nucleotide excision repair (NER) has allowed bacteria to flourish in many different niches around the globe that inflict harsh environmental damage to their genetic material. NER is remarkable because of its diverse substrate repertoire, which differs greatly in chemical composition and structure. Recent advances in structural biology and single-molecule studies have given great insight into the structure and function of NER components. This ensemble of proteins orchestrates faithful removal of toxic DNA lesions through a multistep process. The damaged nucleotide is recognized by dynamic probing of the DNA structure that is then verified and marked for dual incisions followed by excision of the damage and surrounding nucleotides. The opposite DNA strand serves as a template for repair, which is completed after resynthesis and ligation.
Figures




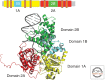

References
-
- Ahn B 2000. A physical interaction of UvrD with nucleotide excision repair protein UvrB. Mol Cells 10: 592–597 - PubMed
-
- Alexandrovich A, Czisch M, Frenkiel TA, Kelly GP, Goosen N, Moolenaar GF, Chowdhry BZ, Sanderson MR, Lane AN 2001. Solution structure, hydrodynamics and thermodynamics of the UvrB C-terminal domain. J Biomol Struct Dyn 19: 219–236 - PubMed
-
- Assenmacher N, Wenig K, Lammens A, Hopfner KP 2006. Structural basis for transcription-coupled repair: The N terminus of Mfd resembles UvrB with degenerate ATPase motifs. J Mol Biol 355: 675–683 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
