Translesion DNA synthesis and mutagenesis in eukaryotes
- PMID: 23457261
- PMCID: PMC3578355
- DOI: 10.1101/cshperspect.a012708
Translesion DNA synthesis and mutagenesis in eukaryotes
Abstract
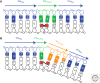
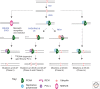
The structural features that enable replicative DNA polymerases to synthesize DNA rapidly and accurately also limit their ability to copy damaged DNA. Direct replication of DNA damage is termed translesion synthesis (TLS), a mechanism conserved from bacteria to mammals and executed by an array of specialized DNA polymerases. This chapter examines how these translesion polymerases replicate damaged DNA and how they are regulated to balance their ability to replicate DNA lesions with the risk of undesirable mutagenesis. It also discusses how TLS is co-opted to increase the diversity of the immunoglobulin gene hypermutation and the contribution it makes to the mutations that sculpt the genome of cancer cells.
Figures


References
-
- Albertella MR, Green CM, Lehmann AR, O’Connor MJ 2005a. A role for polymerase η in the cellular tolerance to cisplatin-induced damage. Cancer Res 65: 9799–9806 - PubMed
-
- Albertella MR, Lau A, O’Connor MJ 2005b. The overexpression of specialized DNA polymerases in cancer. DNA Repair (Amst) 4: 583–593 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
