Curcumin modulates DNA methylation in colorectal cancer cells
- PMID: 23460897
- PMCID: PMC3584082
- DOI: 10.1371/journal.pone.0057709
Curcumin modulates DNA methylation in colorectal cancer cells
Abstract
Aim: Recent evidence suggests that several dietary polyphenols may exert their chemopreventive effect through epigenetic modifications. Curcumin is one of the most widely studied dietary chemopreventive agents for colon cancer prevention, however, its effects on epigenetic alterations, particularly DNA methylation, remain unclear. Using systematic genome-wide approaches, we aimed to elucidate the effect of curcumin on DNA methylation alterations in colorectal cancer cells.
Materials and methods: To evaluate the effect of curcumin on DNA methylation, three CRC cell lines, HCT116, HT29 and RKO, were treated with curcumin. 5-aza-2'-deoxycytidine (5-aza-CdR) and trichostatin A treated cells were used as positive and negative controls for DNA methylation changes, respectively. Methylation status of LINE-1 repeat elements, DNA promoter methylation microarrays and gene expression arrays were used to assess global methylation and gene expression changes. Validation was performed using independent microarrays, quantitative bisulfite pyrosequencing, and qPCR.
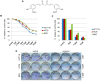
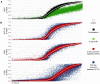
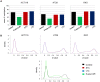
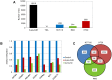
Results: As expected, genome-wide methylation microarrays revealed significant DNA hypomethylation in 5-aza-CdR-treated cells (mean β-values of 0.12), however, non-significant changes in mean β-values were observed in curcumin-treated cells. In comparison to mock-treated cells, curcumin-induced DNA methylation alterations occurred in a time-dependent manner. In contrast to the generalized, non-specific global hypomethylation observed with 5-aza-CdR, curcumin treatment resulted in methylation changes at selected, partially-methylated loci, instead of fully-methylated CpG sites. DNA methylation alterations were supported by corresponding changes in gene expression at both up- and down-regulated genes in various CRC cell lines.
Conclusions: Our data provide previously unrecognized evidence for curcumin-mediated DNA methylation alterations as a potential mechanism of colon cancer chemoprevention. In contrast to non-specific global hypomethylation induced by 5-aza-CdR, curcumin-induced methylation changes occurred only in a subset of partially-methylated genes, which provides additional mechanistic insights into the potent chemopreventive effect of this dietary nutraceutical.
Conflict of interest statement
Figures


References
-
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, et al. (2009) Cancer statistics, 2009. CA Cancer J Clin 59: 225–249. - PubMed
-
- Feinberg AP, Ohlsson R, Henikoff S (2006) The epigenetic progenitor origin of human cancer. Nat Rev Genet 7: 21–33. - PubMed
-
- Feinberg AP (2008) Epigenetics at the epicenter of modern medicine. JAMA 299: 1345–1350. - PubMed
-
- Doll R, Peto R (1981) The causes of cancer: quantitative estimates of avoidable risks of cancer in the United States today. J Natl Cancer Inst 66: 1191–1308. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

