Evolutionary Genomics of Salmonella enterica Subspecies
- PMID: 23462113
- PMCID: PMC3604774
- DOI: 10.1128/mBio.00579-12
Evolutionary Genomics of Salmonella enterica Subspecies
Erratum in
- MBio. 2013;4(2):e00198-13. Bhonagiri-Palsikar, Veena [added]; Hallsworth-Pepin, Kymberlie [added]
Abstract
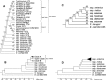
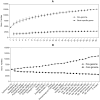
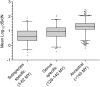
ABSTRACT Six subspecies are currently recognized in Salmonella enterica. Subspecies I (subspecies enterica) is responsible for nearly all infections in humans and warm-blooded animals, while five other subspecies are isolated principally from cold-blooded animals. We sequenced 21 phylogenetically diverse strains, including two representatives from each of the previously unsequenced five subspecies and 11 diverse new strains from S. enterica subspecies enterica, to put this species into an evolutionary perspective. The phylogeny of the subspecies was partly obscured by abundant recombination events between lineages and a relatively short period of time within which subspeciation took place. Nevertheless, a variety of different tree-building methods gave congruent evolutionary tree topologies for subspeciation. A total of 285 gene families were identified that were recruited into subspecies enterica, and most of these are of unknown function. At least 2,807 gene families were identified in one or more of the other subspecies that are not found in subspecies I or Salmonella bongori. Among these gene families were 13 new candidate effectors and 7 new candidate fimbrial clusters. A third complete type III secretion system not present in subspecies enterica (I) isolates was found in both strains of subspecies salamae (II). Some gene families had complex taxonomies, such as the type VI secretion systems, which were recruited from four different lineages in five of six subspecies. Analysis of nonsynonymous-to-synonymous substitution rates indicated that the more-recently acquired regions in S. enterica are undergoing faster fixation rates than the rest of the genome. Recently acquired AT-rich regions, which often encode virulence functions, are under ongoing selection to maintain their high AT content. IMPORTANCE We have sequenced 21 new genomes which encompass the phylogenetic diversity of Salmonella, including strains of the previously unsequenced subspecies arizonae, diarizonae, houtenae, salamae, and indica as well as new diverse strains of subspecies enterica. We have deduced possible evolutionary paths traversed by this very important zoonotic pathogen and identified novel putative virulence factors that are not found in subspecies I. Gene families gained at the time of the evolution of subspecies enterica are of particular interest because they include mechanisms by which this subspecies adapted to warm-blooded hosts.
Figures


References
-
- Chimalizeni Y, Kawaza K, Molyneux E. 2010. The epidemiology and management of non typhoidal salmonella infections. Adv. Exp. Med. Biol. 659:33–46 - PubMed
-
- Frenzen PD, Riggs TL, Buzby JC, Breuer T, Roberts T, Voetsch D, Reddy S. 1999. Salmonella cost estimate updated using FoodNet data. J. Food Saf. 22:10–15
-
- Schlundt J, Toyofuku H, Jansen J, Herbst SA. 2004. Emerging food-borne zoonoses. Rev. Sci. Tech. 23:513–533 - PubMed
-
- Beltran P, Musser JM, Helmuth R, Farmer JJ, III, Frerichs WM, Wachsmuth IK, Ferris K, McWhorter AC, Wells JG, Cravioto A, Selander RK. 1988. Toward a population genetic analysis of Salmonella: genetic diversity and relationships among strains of serotypes S. choleraesuis, S. derby, S. dublin, S. enteritidis, S. heidelberg, S. infantis, S. newport, and S. typhimurium. Proc. Natl. Acad. Sci. U. S. A. 85:7753–7757 - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

