Haemogenic endocardium contributes to transient definitive haematopoiesis
- PMID: 23463007
- PMCID: PMC3612528
- DOI: 10.1038/ncomms2569
Haemogenic endocardium contributes to transient definitive haematopoiesis
Abstract
Haematopoietic cells arise from spatiotemporally restricted domains in the developing embryo. Although studies of non-mammalian animal and in vitro embryonic stem cell models suggest a close relationship among cardiac, endocardial and haematopoietic lineages, it remains unknown whether the mammalian heart tube serves as a haemogenic organ akin to the dorsal aorta. Here we examine the haemogenic activity of the developing endocardium. Mouse heart explants generate myeloid and erythroid colonies in the absence of circulation. Haemogenic activity arises from a subset of endocardial cells in the outflow cushion and atria earlier than in the aorta-gonad-mesonephros region, and is transient and definitive in nature. Interestingly, key cardiac transcription factors, Nkx2-5 and Isl1, are expressed in and required for the haemogenic population of the endocardium. Together, these data suggest that a subset of endocardial/endothelial cells serve as a de novo source for transient definitive haematopoietic progenitors.
Figures
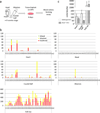
Schematic representation of the colony forming assay from ex vivo organ explant at pre-circulation stages. The heart tube, head, allantois, caudal half (including future AGM region) and yolk sac were dissected at somite stages 1–5, before the formation of effective circulatory loop. Tissues were washed in 3 changes of PBS and cultured on an OP9 feeder layer for 4 days, followed by methylcellulose culture in the presence of hematopoietic growth factors.
Hematopoietic colonies retrieved from various tissues at various somite stages. Each column represents colonies from one tissue. The heart tubes displayed hematopoietic activity whereas the head explants did not. Note the difference in the scale in the yolk sac.
Colonies from Ncx1 mutant embryos that lack heartbeat, showing the hematopoietic activity in the heart tube in the absence of effective heartbeat. Mean±SEM.
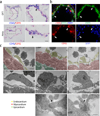
Immunohistochemistry for CD41 (blue) and CD31 (red) in outflow tract (OFT) and atria at E9.5. CD41+ endocardial cells are clustered in the outflow cushion and the dorsal wall of the atria, and also scattered in the entire circumference (arrows). Scale bar = 100µm (lower magnification), 50µm (higher magnification).
Immunofluorescent staining for CD41 (red), CD31 (green) and DAPI (blue) in left atrium at E9.5. Arrows indicate scattered CD31+ endocardial cells co-expressing CD41. Scale bar = 20µm.
Electron microscopic analysis of E10.0–10.5 atrium. Bottom panels show higher magnification of the broken-lined area in upper panels. Left panels indicate regular flat endocardial cells. Center panels are representative images of endocardial cells rounded up. Endothelial cells occasionally protrude into the cardiac lumen (right panels). The bottom panels are higher magnification of adherens junctions in the dotted area in the middle panels (arrows). Scale bar = 2µm (upper panels), 1µm (middle panels) and 100nm (bottom panels). Yellow = endocardial layer. Red = myocardial layer.
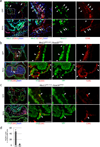
Immunofluorescent staining for Nkx2-5 (green) and CD31 (red) at E9.5. Clusters of Nkx2-5+ cells are found in the endocardium of outflow cushion, atrioventricular cushion, and the dorsal wall of the atria (white arrows). Note that myocardium shows stronger level of Nkx2-5 expression (white arrowheads). Endodermal cells are also positive for Nkx2-5 (left lower panel, open arrowhead). Scale bar = 50µm (lower magnification), 20µm (higher magnification).
Heart section of Nkx2-5IRES-Cre/+; R26YFPR/+ embryo at E10.5 stained for YFP (green) and CD31 (red). Nkx2-5-derived endocardial cells (YFP/CD31 double positive) were clustered in the outflow cushion and the dorsal wall of the atrium, and also found scattered along the perimeter (arrows). This specific distribution pattern corresponds to that of CD41+ cells (Fig. 2a, b). Arrowheads indicate non-Nkx2-5-derived endocardial cells. Scale bar = 100µm (lower magnification), 20µm (higher magnification).
Heart section of Nkx2-5IRES-Cre/+; R26YFPR/+ embryo at E10.5 stained for YFP (green) and CD41 (red). Nkx2-5+ cells give rise to CD41+ cells in the endocardium (YFP/CD41 double positive, arrows). Scale bar = 100µm (lower magnification), 20µm (higher magnification).
Percentage of CD41+ cells in Nkx2-5-derived fraction of endocardium at E9.5. 48% of Nkx2-5-lineage labeled endocardial cells express CD41 whereas 4.4% are CD41-positive in non-Nkx2-5-derived endocardial cells. The graph represents the average of three independent experiments. Mean±SEM.
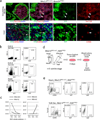
Peripheral blood and liver in Nkx2-5IRES-Cre/+; R26YFPR/+ E10.5 embryo stained for YFP (green) and Ter119 (red). Double positive cells (arrows) indicate circulating erythroids (upper panels) and CD45+ cells repopulating in the liver (lower panels) are positive for YFP. Note that some endodermal cells in the liver are also labeled by YFP reporter. Scale bar = 10µm.
Peripheral blood cells from Nkx2-5IRES-Cre/+; R26YFPR/+ embryos at E10.5, E15.5 and adult stages were analyzed for YFP, CD45 and Ter119 by FACS. Nkx2-5+ cells contribute to 4.0% of Ter119+ cells and 12.0% of CD45+ cells at E10.5, and 2.9% and 0.1%, respectively, at E15.5. No YFP+ blood cells are identified in the adult peripheral circulation.
Expression analyses of β-globins in Nkx2-5-derived peripheral blood at E10.5, 12.5 and 15.5 by qPCR from YFP+ circulating cells of Nkx2-5IRES-Cre/+; R26YFPR/+ embryos. Hbb-bh1 (solid line, left) was first downregulated by E12.5 followed by downregulation of Ey (dashed line, left) by E15.5. Hbb-b1 and b2 were strongly upregulated by E15.5.
Primitive and definitive erythroids were sorted from E15.5 control and Nkx2-5IRES-Cre/+; R26YFPR/+ embryos based on forward and side scatters (top left panels). FACS analyses for Hoechst and YFP revealed that Nkx2-5+ cells contribute to both nucleated primitive erythroids (solid blue line) and enucleated definitive erythroids (solid red line, top right panels). qPCR analysis for β-globins revealed that Nkx2-5-derived primitive and definitive erythroids dominantly express Hbb-Ey and Hbb-b1/b2, respectively (bottom panel).
Schematic representation of the colony forming assay from the explant of Nkx2-5-lineage labeled organs at pre-circulation stages. Tissues were washed in 3 changes of PBS and cultured on OP9 feeder for 4 days, followed by methylcellulose culture in the presence of hematopoietic growth factors.
FACS analysis of the colonies retrieved from Nkx2-5-lineage labeled heart tubes and yolk sacs at pre-circulation stages. Nkx2-5-derived cells constitute 54.0% of Ter119+ cells and 96.4% of CD45+ cells in the colonies from the heart tubes (left), but only 3.0% and 2.2%, respectively, in those from the yolk sac (right).
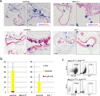
Nkx2-5 mutant heart at E9.5 stained for CD41 (blue) and CD31 (red). CD41+ cells were hardly found in the endocardium of Nkx2-5 mutants. Note that Nkx2-5 mutant also develops known cardiac defects including a single ventricle with poorly developed trabeculae, open atrioventricular canal, and lack of epithelial-mesenchymal transformation in the endocardial cushion. Scale bar = 100µm.
Nkx2-5-derived endocardial cells in Nkx2-5 hetero (left panels) and null (right panels) background. Nkx2-5-driving cells were identified in the cushion endocardium even in the absence of Nkx2-5.
Blood colony formation assay from FACS-purified endocardial cells. No colonies were formed from Nkx2-5 mutant endocardial cells. Data represent average of three independent experiments.
Comparison of the peripheral blood cells derived from Nkx2-5-expressing cells in Nkx2-5-null and its control backgrounds at E10.0. YFP-labeled red blood cells (Ter119+) were found less frequently (0.2%) in Nkx2-5Cre/Cre; R26YFPR/+ embryos compared with control littermates (1.7%), suggesting that expression of Nkx2-5 is required for the hemogenic activity of the Nkx2.5+ endocardial/endothelial cells in vivo.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

