Huntington disease in the South African population occurs on diverse and ethnically distinct genetic haplotypes
- PMID: 23463025
- PMCID: PMC3778359
- DOI: 10.1038/ejhg.2013.2
Huntington disease in the South African population occurs on diverse and ethnically distinct genetic haplotypes
Abstract
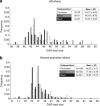
Huntington disease (HD) is a neurodegenerative disorder resulting from the expansion of a CAG trinucleotide repeat in the huntingtin (HTT) gene. Worldwide prevalence varies geographically with the highest figures reported in populations of European ancestry. HD in South Africa has been reported in Caucasian, black and mixed subpopulations, with similar estimated prevalence in the Caucasian and mixed groups and a lower estimate in the black subpopulation. Recent studies have associated specific HTT haplotypes with HD in distinct populations. Expanded HD alleles in Europe occur predominantly on haplogroup A (specifically high-risk variants A1/A2), whereas in East Asian populations, HD alleles are associated with haplogroup C. Whether specific HTT haplotypes associate with HD in black Africans and how these compare with haplotypes found in European and East Asian populations remains unknown. The current study genotyped the HTT region in unaffected individuals and HD patients from each of the South African subpopulations, and haplotypes were constructed. CAG repeat sizes were determined and phased to haplotype. Results indicate that HD alleles from Caucasian and mixed patients are predominantly associated with haplogroup A, signifying a similar European origin for HD. However, in black patients, HD occurs predominantly on haplogroup B, suggesting several distinct origins of the mutation in South Africa. The absence of high-risk variants (A1/A2) in the black subpopulation may also explain the reported low prevalence of HD. Identification of haplotypes associated with HD-expanded alleles is particularly relevant to the development of population-specific therapeutic targets for selective suppression of the expanded HTT transcript.
Figures



References
-
- Walker FO. Huntington's disease. Lancet. 2007;369:218–228. - PubMed
-
- Harper PS. The epidemiology of Huntington's disease. Hum Genet. 1992;89:365–376. - PubMed
-
- Kremer B, Goldberg P, Andrew SE, et al. A worldwide study of the Huntington's disease mutation. The sensitivity and specificity of measuring CAG repeats. New Engl J Med. 1994;330:1401–1406. - PubMed
-
- Pearson CE, Nichol EK, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet. 2005;6:729–742. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

