Relationship between bacterial strain type, host biomarkers, and mortality in Clostridium difficile infection
- PMID: 23463640
- PMCID: PMC3641870
- DOI: 10.1093/cid/cit127
Relationship between bacterial strain type, host biomarkers, and mortality in Clostridium difficile infection
Abstract
Background: Despite substantial interest in biomarkers, their impact on clinical outcomes and variation with bacterial strain has rarely been explored using integrated databases.
Methods: From September 2006 to May 2011, strains isolated from Clostridium difficile toxin enzyme immunoassay (EIA)-positive fecal samples from Oxfordshire, United Kingdom (approximately 600,000 people) underwent multilocus sequence typing. Fourteen-day mortality and levels of 15 baseline biomarkers were compared between consecutive C. difficile infections (CDIs) from different clades/sequence types (STs) and EIA-negative controls using Cox and normal regression adjusted for demographic/clinical factors.
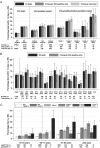
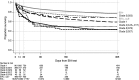
Results: Fourteen-day mortality was 13% in 2222 adults with 2745 EIA-positive samples (median, 78 years) vs 5% in 20,722 adults with 27,550 EIA-negative samples (median, 74 years) (absolute attributable mortality, 7.7%; 95% CI, 6.4%-9.0%). Mortality was highest in clade 5 CDIs (25% [16 of 63]; polymerase chain reaction (PCR) ribotype 078/ST 11), then clade 2 (20% [111 of 560]; 99% PCR ribotype 027/ST 1) versus clade 1 (12% [137 of 1168]; adjusted P < .0001). Within clade 1, 14-day mortality was only 4% (3 of 84) in ST 44 (PCR ribotype 015) (adjusted P = .05 vs other clade 1). Mean baseline neutrophil counts also varied significantly by genotype: 12.4, 11.6, and 9.5 × 10(9) neutrophils/L for clades 5, 2 and 1, respectively, vs 7.0 × 10(9) neutrophils/L in EIA-negative controls (P < .0001) and 7.9 × 10(9) neutrophils/L in ST 44 (P = .08). There were strong associations between C. difficile-type-specific effects on mortality and neutrophil/white cell counts (rho = 0.48), C-reactive-protein (rho = 0.43), eosinophil counts (rho = -0.45), and serum albumin (rho = -0.47). Biomarkers predicted 30%-40% of clade-specific mortality differences.
Conclusions: C. difficile genotype predicts mortality, and excess mortality correlates with genotype-specific changes in biomarkers, strongly implicating inflammatory pathways as a major influence on poor outcome after CDI. PCR ribotype 078/ST 11 (clade 5) leads to severe CDI; thus ongoing surveillance remains essential.
Keywords: C. difficile; biomarkers; mortality; strain-specific variation.
Figures



Comment in
-
Does infection with specific Clostridium difficile strains or clades influence clinical outcome?Clin Infect Dis. 2013 Jun;56(11):1601-3. doi: 10.1093/cid/cit133. Epub 2013 Mar 5. Clin Infect Dis. 2013. PMID: 23463642 No abstract available.
-
Reply to Walk et al.Clin Infect Dis. 2013 Aug;57(4):626-7. doi: 10.1093/cid/cit312. Epub 2013 May 8. Clin Infect Dis. 2013. PMID: 23658434 No abstract available.
-
Understanding increased mortality in Clostridium difficile-infected older adults.Clin Infect Dis. 2013 Aug;57(4):625-6. doi: 10.1093/cid/cit310. Epub 2013 May 8. Clin Infect Dis. 2013. PMID: 23658435 Free PMC article. No abstract available.
References
-
- Loo VG, Poirier L, Miller MA, et al. A predominantly clonal multi-institutional outbreak of Clostridium difficile–associated diarrhea with high morbidity and mortality. N Engl J Med. 2005;353:2442–9. - PubMed
-
- McDonald LC, Killgore GE, Thompson A, et al. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med. 2005;353:2433–41. - PubMed
-
- Miller M, Gravel D, Mulvey M, et al. Health care×associated Clostridium difficile infection in Canada: patient age and infecting strain type are highly predictive of severe outcome and mortality. Clin Infect Dis. 2010;50:194–201. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

