Nutrient signaling to mTOR and cell growth
- PMID: 23465396
- PMCID: PMC3634910
- DOI: 10.1016/j.tibs.2013.01.004
Nutrient signaling to mTOR and cell growth
Abstract
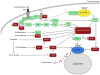
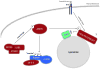
The mammalian target of rapamycin (mTOR) is a conserved protein kinase involved in a multitude of cellular processes including cell growth. Increased mTOR activation is observed in multiple human cancers and inhibition of mTOR has proven efficacious in numerous clinical trials. mTOR comprises two complexes, termed mTORC1 and mTORC2. Both complexes respond to growth factors, whereas only mTORC1 is controlled by nutrients, such as glucose and amino acids. Since the discovery of mTOR, extensive studies have intricately detailed the molecular mechanisms by which mTORC1 is regulated. Somewhat paradoxically, amino acid (AA)-induced mTORC1 activation -arguably the most essential stimulus leading to mTORC1 activation - is the least understood. Here we review the current knowledge of nutrient-dependent regulation of mTORC1.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures

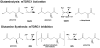
References
-
- Heitman J, et al. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. - PubMed
-
- Kunz J, et al. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–596. - PubMed
-
- Sabatini DM, et al. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. - PubMed
-
- Sabers CJ, et al. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J Biol Chem. 1995;270:815–822. - PubMed
-
- Brown EJ, et al. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

