High-resolution SAR11 ecotype dynamics at the Bermuda Atlantic Time-series Study site by phylogenetic placement of pyrosequences
- PMID: 23466704
- PMCID: PMC3695298
- DOI: 10.1038/ismej.2013.32
High-resolution SAR11 ecotype dynamics at the Bermuda Atlantic Time-series Study site by phylogenetic placement of pyrosequences
Abstract
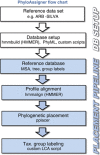
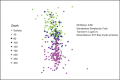
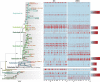
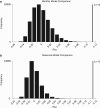
Advances in next-generation sequencing technologies are providing longer nucleotide sequence reads that contain more information about phylogenetic relationships. We sought to use this information to understand the evolution and ecology of bacterioplankton at our long-term study site in the Western Sargasso Sea. A bioinformatics pipeline called PhyloAssigner was developed to align pyrosequencing reads to a reference multiple sequence alignment of 16S ribosomal RNA (rRNA) genes and assign them phylogenetic positions in a reference tree using a maximum likelihood algorithm. Here, we used this pipeline to investigate the ecologically important SAR11 clade of Alphaproteobacteria. A combined set of 2.7 million pyrosequencing reads from the 16S rRNA V1-V2 regions, representing 9 years at the Bermuda Atlantic Time-series Study (BATS) site, was quality checked and parsed into a comprehensive bacterial tree, yielding 929 036 Alphaproteobacteria reads. Phylogenetic structure within the SAR11 clade was linked to seasonally recurring spatiotemporal patterns. This analysis resolved four new SAR11 ecotypes in addition to five others that had been described previously at BATS. The data support a conclusion reached previously that the SAR11 clade diversified by subdivision of niche space in the ocean water column, but the new data reveal a more complex pattern in which deep branches of the clade diversified repeatedly across depth strata and seasonal regimes. The new data also revealed the presence of an unrecognized clade of Alphaproteobacteria, here named SMA-1 (Sargasso Mesopelagic Alphaproteobacteria, group 1), in the upper mesopelagic zone. The high-resolution phylogenetic analyses performed herein highlight significant, previously unknown, patterns of evolutionary diversification, within perhaps the most widely distributed heterotrophic marine bacterial clade, and strongly links to ecosystem regimes.
Figures


References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

