The BEN domain is a novel sequence-specific DNA-binding domain conserved in neural transcriptional repressors
- PMID: 23468431
- PMCID: PMC3613608
- DOI: 10.1101/gad.213314.113
The BEN domain is a novel sequence-specific DNA-binding domain conserved in neural transcriptional repressors
Abstract
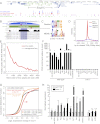
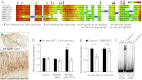
We recently reported that Drosophila Insensitive (Insv) promotes sensory organ development and has activity as a nuclear corepressor for the Notch transcription factor Suppressor of Hairless [Su(H)]. Insv lacks domains of known biochemical function but contains a single BEN domain (i.e., a "BEN-solo" protein). Our chromatin immunoprecipitation (ChIP) sequencing (ChIP-seq) analysis confirmed binding of Insensitive to Su(H) target genes in the Enhancer of split gene complex [E(spl)-C]; however, de novo motif analysis revealed a novel site strongly enriched in Insv peaks (TCYAATHRGAA). We validate binding of endogenous Insv to genomic regions bearing such sites, whose associated genes are enriched for neural functions and are functionally repressed by Insv. Unexpectedly, we found that the Insv BEN domain binds specifically to this sequence motif and that Insv directly regulates transcription via this motif. We determined the crystal structure of the BEN-DNA target complex, revealing homodimeric binding of the BEN domain and extensive nucleotide contacts via α helices and a C-terminal loop. Point mutations in key DNA-contacting residues severely impair DNA binding in vitro and capacity for transcriptional regulation in vivo. We further demonstrate DNA-binding and repression activities by the mammalian neural BEN-solo protein BEND5. Altogether, we define novel DNA-binding activity in a conserved family of transcriptional repressors, opening a molecular window on this extensive gene family.
Figures




References
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC 2002. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 - PubMed
-
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
