MSH3 polymorphisms and protein levels affect CAG repeat instability in Huntington's disease mice
- PMID: 23468640
- PMCID: PMC3585117
- DOI: 10.1371/journal.pgen.1003280
MSH3 polymorphisms and protein levels affect CAG repeat instability in Huntington's disease mice
Abstract
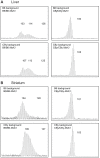
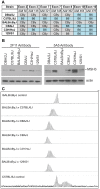
Expansions of trinucleotide CAG/CTG repeats in somatic tissues are thought to contribute to ongoing disease progression through an affected individual's life with Huntington's disease or myotonic dystrophy. Broad ranges of repeat instability arise between individuals with expanded repeats, suggesting the existence of modifiers of repeat instability. Mice with expanded CAG/CTG repeats show variable levels of instability depending upon mouse strain. However, to date the genetic modifiers underlying these differences have not been identified. We show that in liver and striatum the R6/1 Huntington's disease (HD) (CAG)∼100 transgene, when present in a congenic C57BL/6J (B6) background, incurred expansion-biased repeat mutations, whereas the repeat was stable in a congenic BALB/cByJ (CBy) background. Reciprocal congenic mice revealed the Msh3 gene as the determinant for the differences in repeat instability. Expansion bias was observed in congenic mice homozygous for the B6 Msh3 gene on a CBy background, while the CAG tract was stabilized in congenics homozygous for the CBy Msh3 gene on a B6 background. The CAG stabilization was as dramatic as genetic deficiency of Msh2. The B6 and CBy Msh3 genes had identical promoters but differed in coding regions and showed strikingly different protein levels. B6 MSH3 variant protein is highly expressed and associated with CAG expansions, while the CBy MSH3 variant protein is expressed at barely detectable levels, associating with CAG stability. The DHFR protein, which is divergently transcribed from a promoter shared by the Msh3 gene, did not show varied levels between mouse strains. Thus, naturally occurring MSH3 protein polymorphisms are modifiers of CAG repeat instability, likely through variable MSH3 protein stability. Since evidence supports that somatic CAG instability is a modifier and predictor of disease, our data are consistent with the hypothesis that variable levels of CAG instability associated with polymorphisms of DNA repair genes may have prognostic implications for various repeat-associated diseases.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Rosenblatt A, Kumar BV, Mo A, Welsh CS, Margolis RL, et al. (2012) Age, CAG repeat length, and clinical progression in Huntington's disease. Mov Disord 27: 272–276. - PubMed
-
- Telenius H, Kremer B, Goldberg YP, Theilmann J, Andrew SE, et al. (1994) Somatic and gonadal mosaicism of the Huntington disease gene CAG repeat in brain and sperm. Nat Genet 6: 409–414. - PubMed
-
- De Rooij KE, De Koning Gans PA, Roos RA, Van Ommen GJ, Den Dunnen JT (1995) Somatic expansion of the (CAG)n repeat in Huntington disease brains. Hum Genet 95: 270–274. - PubMed
-
- Kennedy L, Evans E, Chen CM, Craven L, Detloff PJ, et al. (2003) Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum Mol Genet 12: 3359–3367. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

