MicroRNA-3148 modulates allelic expression of toll-like receptor 7 variant associated with systemic lupus erythematosus
- PMID: 23468661
- PMCID: PMC3585142
- DOI: 10.1371/journal.pgen.1003336
MicroRNA-3148 modulates allelic expression of toll-like receptor 7 variant associated with systemic lupus erythematosus
Abstract
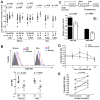
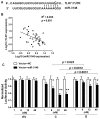
We previously reported that the G allele of rs3853839 at 3'untranslated region (UTR) of Toll-like receptor 7 (TLR7) was associated with elevated transcript expression and increased risk for systemic lupus erythematosus (SLE) in 9,274 Eastern Asians [P = 6.5×10(-10), odds ratio (OR) (95%CI) = 1.27 (1.17-1.36)]. Here, we conducted trans-ancestral fine-mapping in 13,339 subjects including European Americans, African Americans, and Amerindian/Hispanics and confirmed rs3853839 as the only variant within the TLR7-TLR8 region exhibiting consistent and independent association with SLE (Pmeta = 7.5×10(-11), OR = 1.24 [1.18-1.34]). The risk G allele was associated with significantly increased levels of TLR7 mRNA and protein in peripheral blood mononuclear cells (PBMCs) and elevated luciferase activity of reporter gene in transfected cells. TLR7 3'UTR sequence bearing the non-risk C allele of rs3853839 matches a predicted binding site of microRNA-3148 (miR-3148), suggesting that this microRNA may regulate TLR7 expression. Indeed, miR-3148 levels were inversely correlated with TLR7 transcript levels in PBMCs from SLE patients and controls (R(2) = 0.255, P = 0.001). Overexpression of miR-3148 in HEK-293 cells led to significant dose-dependent decrease in luciferase activity for construct driven by TLR7 3'UTR segment bearing the C allele (P = 0.0003). Compared with the G-allele construct, the C-allele construct showed greater than two-fold reduction of luciferase activity in the presence of miR-3148. Reduced modulation by miR-3148 conferred slower degradation of the risk G-allele containing TLR7 transcripts, resulting in elevated levels of gene products. These data establish rs3853839 of TLR7 as a shared risk variant of SLE in 22,613 subjects of Asian, EA, AA, and Amerindian/Hispanic ancestries (Pmeta = 2.0×10(-19), OR = 1.25 [1.20-1.32]), which confers allelic effect on transcript turnover via differential binding to the epigenetic factor miR-3148.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Kontaki E, Boumpas DT (2010) Innate immunity in systemic lupus erythematosus: sensing endogenous nucleic acids. J Autoimmun 35: 206–211. - PubMed
-
- Bronson PG, Chaivorapol C, Ortmann W, Behrens TW, Graham RR (2012) The genetics of type I interferon in systemic lupus erythematosus. Curr Opin Immunol [Epub ahead of print]. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UL1 RR025005/RR/NCRR NIH HHS/United States
- UL1 TR000154/TR/NCATS NIH HHS/United States
- R01AI063274/AI/NIAID NIH HHS/United States
- K24 AI078004/AI/NIAID NIH HHS/United States
- P60 AR030692/AR/NIAMS NIH HHS/United States
- R21AI070304/AI/NIAID NIH HHS/United States
- UL1RR025005/RR/NCRR NIH HHS/United States
- UL1 RR033176/RR/NCRR NIH HHS/United States
- P60 AR053308/AR/NIAMS NIH HHS/United States
- R01 AI063274/AI/NIAID NIH HHS/United States
- M01 RR000079/RR/NCRR NIH HHS/United States
- P01AR49084/AR/NIAMS NIH HHS/United States
- UL1RR033176/RR/NCRR NIH HHS/United States
- UL1TR000124/TR/NCATS NIH HHS/United States
- U54 GM104938/GM/NIGMS NIH HHS/United States
- UL1TR000165/TR/NCATS NIH HHS/United States
- M01RR-00079/RR/NCRR NIH HHS/United States
- P20 RR020143/RR/NCRR NIH HHS/United States
- P30 GM103510/GM/NIGMS NIH HHS/United States
- U19AI082714/AI/NIAID NIH HHS/United States
- U19 AI082714/AI/NIAID NIH HHS/United States
- UL1 TR000150/TR/NCATS NIH HHS/United States
- R01AR057172/AR/NIAMS NIH HHS/United States
- UL1RR025741/RR/NCRR NIH HHS/United States
- P20RR020143/RR/NCRR NIH HHS/United States
- P60AR049459/AR/NIAMS NIH HHS/United States
- U01AI090909/AI/NIAID NIH HHS/United States
- P30 AR053483/AR/NIAMS NIH HHS/United States
- UL1 TR000165/TR/NCATS NIH HHS/United States
- UL1 RR024999/RR/NCRR NIH HHS/United States
- R01AR043274/AR/NIAMS NIH HHS/United States
- P01AI083194/AI/NIAID NIH HHS/United States
- K08 AI083790/AI/NIAID NIH HHS/United States
- RC2AR058959/AR/NIAMS NIH HHS/United States
- P01AR052915/AR/NIAMS NIH HHS/United States
- RC1AR058621/AR/NIAMS NIH HHS/United States
- U01 AI090909/AI/NIAID NIH HHS/United States
- UL1 RR025741/RR/NCRR NIH HHS/United States
- R37AI024717/AI/NIAID NIH HHS/United States
- UL1 TR000124/TR/NCATS NIH HHS/United States
- N01 AR062277/AR/NIAMS NIH HHS/United States
- RC1 AR058621/AR/NIAMS NIH HHS/United States
- P30 AR048311/AR/NIAMS NIH HHS/United States
- P30 CA046934/CA/NCI NIH HHS/United States
- R01 AR043814/AR/NIAMS NIH HHS/United States
- P60 AR049459/AR/NIAMS NIH HHS/United States
- R37 AI024717/AI/NIAID NIH HHS/United States
- R01 AR042460/AR/NIAMS NIH HHS/United States
- R01 AR057172/AR/NIAMS NIH HHS/United States
- R01AR051545-01A2/AR/NIAMS NIH HHS/United States
- R01AR33062/AR/NIAMS NIH HHS/United States
- U01 AI101934/AI/NIAID NIH HHS/United States
- R01 AR033062/AR/NIAMS NIH HHS/United States
- R01CA141700/CA/NCI NIH HHS/United States
- K24AR002138/AR/NIAMS NIH HHS/United States
- UL1 RR025014/RR/NCRR NIH HHS/United States
- UL1 RR029882/RR/NCRR NIH HHS/United States
- L30 AI071651/AI/NIAID NIH HHS/United States
- R01AR43727/AR/NIAMS NIH HHS/United States
- R01 AR043727/AR/NIAMS NIH HHS/United States
- R01 CA141700/CA/NCI NIH HHS/United States
- U01AI101934/AI/NIAID NIH HHS/United States
- P30GM103510/GM/NIGMS NIH HHS/United States
- K24 AR002138/AR/NIAMS NIH HHS/United States
- L30AI071651/AI/NIAID NIH HHS/United States
- R01AR042460/AR/NIAMS NIH HHS/United States
- UL1RR025014-02/RR/NCRR NIH HHS/United States
- P60AR053308/AR/NIAMS NIH HHS/United States
- UL1RR024999/RR/NCRR NIH HHS/United States
- R01AR043814/AR/NIAMS NIH HHS/United States
- R21 AI070304/AI/NIAID NIH HHS/United States
- RC2 AR058959/AR/NIAMS NIH HHS/United States
- P01 AI083194/AI/NIAID NIH HHS/United States
- P01 AR052915/AR/NIAMS NIH HHS/United States
- P01 AR049084/AR/NIAMS NIH HHS/United States
- R01 AR051545/AR/NIAMS NIH HHS/United States
- P30AR053483/AR/NIAMS NIH HHS/United States
- P60AR30692/AR/NIAMS NIH HHS/United States
- UL1RR029882/RR/NCRR NIH HHS/United States
- R01 AR043274/AR/NIAMS NIH HHS/United States
- P60 AR062755/AR/NIAMS NIH HHS/United States
- K08AI083790/AI/NIAID NIH HHS/United States
- P30AR48311/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

