De novo assembly, gene annotation and marker development using Illumina paired-end transcriptome sequences in celery (Apium graveolens L.)
- PMID: 23469050
- PMCID: PMC3585167
- DOI: 10.1371/journal.pone.0057686
De novo assembly, gene annotation and marker development using Illumina paired-end transcriptome sequences in celery (Apium graveolens L.)
Abstract
Background: Celery is an increasing popular vegetable species, but limited transcriptome and genomic data hinder the research to it. In addition, a lack of celery molecular markers limits the process of molecular genetic breeding. High-throughput transcriptome sequencing is an efficient method to generate a large transcriptome sequence dataset for gene discovery, molecular marker development and marker-assisted selection breeding.
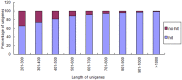
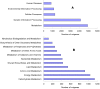
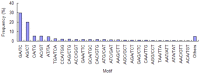
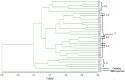
Principal findings: Celery transcriptomes from four tissues were sequenced using Illumina paired-end sequencing technology. De novo assembling was performed to generate a collection of 42,280 unigenes (average length of 502.6 bp) that represent the first transcriptome of the species. 78.43% and 48.93% of the unigenes had significant similarity with proteins in the National Center for Biotechnology Information (NCBI) non-redundant protein database (Nr) and Swiss-Prot database respectively, and 10,473 (24.77%) unigenes were assigned to Clusters of Orthologous Groups (COG). 21,126 (49.97%) unigenes harboring Interpro domains were annotated, in which 15,409 (36.45%) were assigned to Gene Ontology(GO) categories. Additionally, 7,478 unigenes were mapped onto 228 pathways using the Kyoto Encyclopedia of Genes and Genomes Pathway database (KEGG). Large numbers of simple sequence repeats (SSRs) were indentified, and then the rate of successful amplication and polymorphism were investigated among 31 celery accessions.
Conclusions: This study demonstrates the feasibility of generating a large scale of sequence information by Illumina paired-end sequencing and efficient assembling. Our results provide a valuable resource for celery research. The developed molecular markers are the foundation of further genetic linkage analysis and gene localization, and they will be essential to accelerate the process of breeding.
Conflict of interest statement
Figures




References
-
- Muminovic J, Melchinger AE, Lubberstedt T (2004) Prospects for celeriac (Apium graveolens var. rapaceum) improvement by using genetic resources of Apium, as determined by AFLP markers and morphological characterization. Plant Genetic Resources 2: 189–198.
-
- Sampson S (2006) Chinese celery has robust taste. Toronto Star (Canada) ISSN:0319–0781.
-
- Song XJ, Wang YT (2008) Research Progress in the Medicinal Function of Celery. Journal of Anhui Agricultural Sciences 36: 6360–6361, 6395.
-
- TANG F, GUO J, ZHANG J, LI J, SU M (2007) Study on Hypotensive and Vasodilatory Effects of Celery Juice. Food Science 28: 322–325.
-
- Arus P, Ortan T (1984) Inheritance patterns and linkage relationships of eight genes of celery. Journal of Heredity 75: 11–14.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

