In silico structural and functional characterization of the RSUME splice variants
- PMID: 23469069
- PMCID: PMC3585135
- DOI: 10.1371/journal.pone.0057795
In silico structural and functional characterization of the RSUME splice variants
Abstract
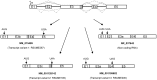
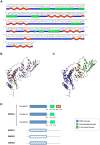
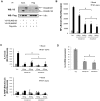
RSUME (RWD-containing SUMO Enhancer) is a small protein that increases SUMO conjugation to proteins. To date, four splice variants that codify three RSUME isoforms have been described, which differ in their C-terminal end. Comparing the structure of the RSUME isoforms we found that, in addition to the previously described RWD domain in the N-terminal, all these RSUME variants also contain an intermediate domain. Only the longest RSUME isoform presents a C-terminal domain that is absent in the others. Given these differences, we used the shortest and longest RSUME variants for comparative studies. We found that the C-terminal domain is dispensable for the SUMO-conjugation enhancer properties of RSUME. We also demonstrate that these two RSUME variants are equally induced by hypoxia. The NF-κB signaling pathway is inhibited and the HIF-1 pathway is increased more efficiently by the longest RSUME, by means of a greater physical interaction of RSUME267 with the target proteins. In addition, the mRNA and protein levels of these isoforms differ in human glioma samples; while the shortest RSUME isoform is expressed in all the tumors analyzed, the longest variant is expressed in most but not all of them. The results presented here show a degree of redundancy of the RSUME variants on the SUMO pathway. However, the increased inhibition conferred by RSUME267 over the NF-κB signaling pathway, the increased activation over the HIF-1 pathway and the different expression of the RSUME isoforms suggest specific roles for each RSUME isoform which may be relevant in certain types of brain tumors that express RSUME, like human pituitary adenomas and gliomas.
Conflict of interest statement
Figures




References
-
- Carbia-Nagashima A, Gerez J, Perez-Castro C, Paez-Pereda M, Silberstein S, et al. (2007) RSUME, a small RWD-containing protein, enhances SUMO conjugation and stabilizes HIF-1alpha during hypoxia. Cell 131: 309–323. - PubMed
-
- Castro CP, Giacomini D, Nagashima AC, Onofri C, Graciarena M, et al. (2003) Reduced expression of the cytokine transducer gp130 inhibits hormone secretion, cell growth, and tumor development of pituitary lactosomatotrophic GH3 cells. Endocrinology 144: 693–700. - PubMed
-
- Frischmeyer PA, Dietz HC (1999) Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet 8: 1893–1900. - PubMed
-
- Hay RT (2005) SUMO: a history of modification. Mol Cell 18: 1–12. - PubMed
-
- Geiss-Friedlander R, Melchior F (2007) Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 8: 947–956. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

