Spatiotemporal dynamics of early DNA damage response proteins on complex DNA lesions
- PMID: 23469115
- PMCID: PMC3582506
- DOI: 10.1371/journal.pone.0057953
Spatiotemporal dynamics of early DNA damage response proteins on complex DNA lesions
Abstract
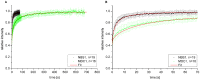
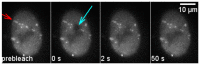
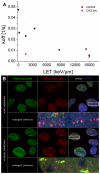
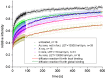
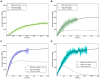
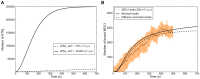
The response of cells to ionizing radiation-induced DNA double-strand breaks (DSB) is determined by the activation of multiple pathways aimed at repairing the injury and maintaining genomic integrity. Densely ionizing radiation induces complex damage consisting of different types of DNA lesions in close proximity that are difficult to repair and may promote carcinogenesis. Little is known about the dynamic behavior of repair proteins on complex lesions. In this study we use live-cell imaging for the spatio-temporal characterization of early protein interactions at damage sites of increasing complexity. Beamline microscopy was used to image living cells expressing fluorescently-tagged proteins during and immediately after charged particle irradiation to reveal protein accumulation at damaged sites in real time. Information on the mobility and binding rates of the recruited proteins was obtained from fluorescence recovery after photobleaching (FRAP). Recruitment of the DNA damage sensor protein NBS1 accelerates with increasing lesion density and saturates at very high damage levels. FRAP measurements revealed two different binding modalities of NBS1 to damage sites and a direct impact of lesion complexity on the binding. Faster recruitment with increasing lesion complexity was also observed for the mediator MDC1, but mobility was limited at very high damage densities due to nuclear-wide binding. We constructed a minimal computer model of the initial response to DSB based on known protein interactions only. By fitting all measured data using the same set of parameters, we can reproduce the experimentally characterized steps of the DNA damage response over a wide range of damage densities. The model suggests that the influence of increasing lesion density accelerating NBS1 recruitment is only dependent on the different binding modes of NBS1, directly to DSB and to the surrounding chromatin via MDC1. This elucidates an impact of damage clustering on repair without the need of invoking extra processing steps.
Conflict of interest statement
Figures






References
-
- Christmann M, Tomicic MT, Roos WP, Kaina B (2003) Mechanisms of human dna repair: an update. Toxicology 193: 3–34. - PubMed
-
- Difilippantonio S, Nussenzweig A (2007) The nbs1-atm connection revisited. Cell Cycle (Georgetown, Tex) 6: 2366–2370. - PubMed
-
- Petrini JHJ, Stracker TH (2003) The cellular response to dna double-strand breaks: defining the sensors and mediators. Trends in Cell Biology 13: 458–462. - PubMed
-
- Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, et al. (2002) The rad50 zinc-hook is a structure joining mre11 complexes in dna recombination and repair. Nature 418: 562–566. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

