TagGD: fast and accurate software for DNA Tag generation and demultiplexing
- PMID: 23469199
- PMCID: PMC3587622
- DOI: 10.1371/journal.pone.0057521
TagGD: fast and accurate software for DNA Tag generation and demultiplexing
Abstract
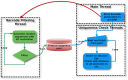
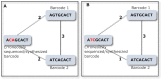
Multiplexing is of vital importance for utilizing the full potential of next generation sequencing technologies. We here report TagGD (DNA-based Tag Generator and Demultiplexor), a fully-customisable, fast and accurate software package that can generate thousands of barcodes satisfying user-defined constraints and can guarantee full demultiplexing accuracy. The barcodes are designed to minimise their interference with the experiment. Insertion, deletion and substitution events are considered when designing and demultiplexing barcodes. 20,000 barcodes of length 18 were designed in 5 minutes and 2 million barcoded Illumina HiSeq-like reads generated with an error rate of 2% were demultiplexed with full accuracy in 5 minutes. We believe that our software meets a central demand in the current high-throughput biology and can be utilised in any field with ample sample abundance. The software is available on GitHub (https://github.com/pelinakan/UBD.git).
Conflict of interest statement
Figures
References
-
- Stahl PL, Lundeberg J (2012) Toward the single-hour high-quality genome. Annual review of biochemistry 81: 359–378. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

