Solid state NMR strategy for characterizing native membrane protein structures
- PMID: 23470103
- PMCID: PMC3715573
- DOI: 10.1021/ar3003442
Solid state NMR strategy for characterizing native membrane protein structures
Abstract
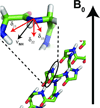
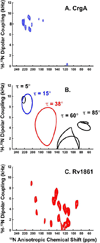
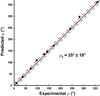
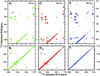
Unlike water soluble proteins, the structures of helical transmembrane proteins depend on a very complex environment. These proteins sit in the midst of dramatic electrical and chemical gradients and are often subject to variations in the lateral pressure profile, order parameters, dielectric constant, and other properties. Solid state NMR is a collection of tools that can characterize high resolution membrane protein structure in this environment. Indeed, prior work has shown that this complex environment significantly influences transmembrane protein structure. Therefore, it is important to characterize such structures under conditions that closely resemble its native environment. Researchers have used two approaches to gain protein structural restraints via solid state NMR spectroscopy. The more traditional approach uses magic angle sample spinning to generate isotropic chemical shifts, much like solution NMR. As with solution NMR, researchers can analyze the backbone chemical shifts to obtain torsional restraints. They can also examine nuclear spin interactions between nearby atoms to obtain distances between atomic sites. Unfortunately, for membrane proteins in lipid preparations, the spectral resolution is not adequate to obtain complete resonance assignments. Researchers have developed another approach for gaining structural restraints from membrane proteins: the use of uniformly oriented lipid bilayers, which provides a method for obtaining high resolution orientational restraints. When the bilayers are aligned with respect to the magnetic field of the NMR spectrometer, researchers can obtain orientational restraints in which atomic sites in the protein are restrained relative to the alignment axis. However, this approach does not allow researchers to determine the relative packing between helices. By combining the two approaches, we can take advantage of the information acquired from each technique to minimize the challenges and maximize the quality of the structural results. By combining the distance, torsional, and orientational restraints, we can characterize high resolution membrane protein structure in native-like lipid bilayer environments.
Figures



References
-
- Page RC, Li C, Hu J, Gao FP, Cross TA. Lipid bilayers: an essential environment for the understanding of membrane proteins. Magn Reson Chem. 2007;45:S2–S11. - PubMed
-
- Ketchem RR, Roux B, Cross TA. High-Resolution Polypeptide Structure in a Lamellar Phase Lipid Environment from Solid-State NMR Derived Orientational Constraints. Structure. 1997;5:1655–1669. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

