Population genetic tools for dissecting innate immunity in humans
- PMID: 23470320
- PMCID: PMC4015519
- DOI: 10.1038/nri3421
Population genetic tools for dissecting innate immunity in humans
Abstract
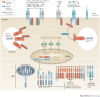
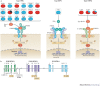
Innate immunity involves direct interactions between the host and microorganisms, both pathogenic and symbiotic, so natural selection is expected to strongly influence genes involved in these processes. Population genetics investigates the impact of past natural selection events on the genome of present-day human populations, and it complements immunological as well as clinical and epidemiological genetic studies. Recent data show that the impact of selection on the different families of innate immune receptors and their downstream signalling molecules varies considerably. This Review discusses these findings and highlights how they help to delineate the relative functional importance of innate immune pathways, which can range from being essential to being redundant.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

