A comparison of dense transposon insertion libraries in the Salmonella serovars Typhi and Typhimurium
- PMID: 23470992
- PMCID: PMC3632133
- DOI: 10.1093/nar/gkt148
A comparison of dense transposon insertion libraries in the Salmonella serovars Typhi and Typhimurium
Abstract
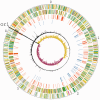
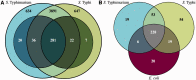
Salmonella Typhi and Typhimurium diverged only ∼50 000 years ago, yet have very different host ranges and pathogenicity. Despite the availability of multiple whole-genome sequences, the genetic differences that have driven these changes in phenotype are only beginning to be understood. In this study, we use transposon-directed insertion-site sequencing to probe differences in gene requirements for competitive growth in rich media between these two closely related serovars. We identify a conserved core of 281 genes that are required for growth in both serovars, 228 of which are essential in Escherichia coli. We are able to identify active prophage elements through the requirement for their repressors. We also find distinct differences in requirements for genes involved in cell surface structure biogenesis and iron utilization. Finally, we demonstrate that transposon-directed insertion-site sequencing is not only applicable to the protein-coding content of the cell but also has sufficient resolution to generate hypotheses regarding the functions of non-coding RNAs (ncRNAs) as well. We are able to assign probable functions to a number of cis-regulatory ncRNA elements, as well as to infer likely differences in trans-acting ncRNA regulatory networks.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

