Comparative genomics of defense systems in archaea and bacteria
- PMID: 23470997
- PMCID: PMC3632139
- DOI: 10.1093/nar/gkt157
Comparative genomics of defense systems in archaea and bacteria
Abstract
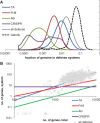
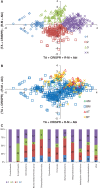
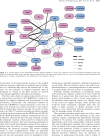
Our knowledge of prokaryotic defense systems has vastly expanded as the result of comparative genomic analysis, followed by experimental validation. This expansion is both quantitative, including the discovery of diverse new examples of known types of defense systems, such as restriction-modification or toxin-antitoxin systems, and qualitative, including the discovery of fundamentally new defense mechanisms, such as the CRISPR-Cas immunity system. Large-scale statistical analysis reveals that the distribution of different defense systems in bacterial and archaeal taxa is non-uniform, with four groups of organisms distinguishable with respect to the overall abundance and the balance between specific types of defense systems. The genes encoding defense system components in bacterial and archaea typically cluster in defense islands. In addition to genes encoding known defense systems, these islands contain numerous uncharacterized genes, which are candidates for new types of defense systems. The tight association of the genes encoding immunity systems and dormancy- or cell death-inducing defense systems in prokaryotic genomes suggests that these two major types of defense are functionally coupled, providing for effective protection at the population level.
Figures



References
-
- Forterre P, Prangishvili D. The great billion-year war between ribosome- and capsid-encoding organisms (cells and viruses) as the major source of evolutionary novelties. Ann. NY Acad. Sci. 2009;1178:65–77. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

