A high-throughput and quantitative method to assess the mutagenic potential of translesion DNA synthesis
- PMID: 23470999
- PMCID: PMC3632128
- DOI: 10.1093/nar/gkt141
A high-throughput and quantitative method to assess the mutagenic potential of translesion DNA synthesis
Abstract
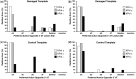
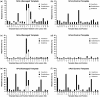
Cellular genomes are constantly damaged by endogenous and exogenous agents that covalently and structurally modify DNA to produce DNA lesions. Although most lesions are mended by various DNA repair pathways in vivo, a significant number of damage sites persist during genomic replication. Our understanding of the mutagenic outcomes derived from these unrepaired DNA lesions has been hindered by the low throughput of existing sequencing methods. Therefore, we have developed a cost-effective high-throughput short oligonucleotide sequencing assay that uses next-generation DNA sequencing technology for the assessment of the mutagenic profiles of translesion DNA synthesis catalyzed by any error-prone DNA polymerase. The vast amount of sequencing data produced were aligned and quantified by using our novel software. As an example, the high-throughput short oligonucleotide sequencing assay was used to analyze the types and frequencies of mutations upstream, downstream and at a site-specifically placed cis-syn thymidine-thymidine dimer generated individually by three lesion-bypass human Y-family DNA polymerases.
Figures

References
-
- Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, et al. The Y-family of DNA polymerases. Mol. Cell. 2001;8:7–8. - PubMed
-
- You YH, Lee DH, Yoon JH, Nakajima S, Yasui A, Pfeifer GP. Cyclobutane pyrimidine dimers are responsible for the vast majority of mutations induced by UVB irradiation in mammalian cells. J. Biol. Chem. 2001;276:44688–44694. - PubMed
-
- Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase eta. J. Biol. Chem. 2000;275:7447–7450. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

