Stability, delivery and functions of human sperm RNAs at fertilization
- PMID: 23471003
- PMCID: PMC3627604
- DOI: 10.1093/nar/gkt132
Stability, delivery and functions of human sperm RNAs at fertilization
Abstract
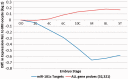
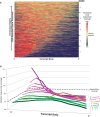
Increasing attention has focused on the significance of RNA in sperm, in light of its contribution to the birth and long-term health of a child, role in sperm function and diagnostic potential. As the composition of sperm RNA is in flux, assigning specific roles to individual RNAs presents a significant challenge. For the first time RNA-seq was used to characterize the population of coding and non-coding transcripts in human sperm. Examining RNA representation as a function of multiple methods of library preparation revealed unique features indicative of very specific and stage-dependent maturation and regulation of sperm RNA, illuminating their various transitional roles. Correlation of sperm transcript abundance with epigenetic marks suggested roles for these elements in the pre- and post-fertilization genome. Several classes of non-coding RNAs including lncRNAs, CARs, pri-miRNAs, novel elements and mRNAs have been identified which, based on factors including relative abundance, integrity in sperm, available knockout data of embryonic effect and presence or absence in the unfertilized human oocyte, are likely to be essential male factors critical to early post-fertilization development. The diverse and unique attributes of sperm transcripts that were revealed provides the first detailed analysis of the biology and anticipated clinical significance of spermatozoal RNAs.
Figures



References
-
- Krawetz SA. Paternal contribution: new insights and future challenges. Nat. Rev. 2005;6:633–642. - PubMed
-
- Pessot CA, Brito M, Figueroa J, Concha II, Yanez A, Burzio LO. Presence of RNA in the sperm nucleus. Biochem. Biophys. Res. Commun. 1989;158:272–278. - PubMed
-
- Sutovsky P, Moreno RD, Ramalho-Santos J, Dominko T, Simerly C, Schatten G. Ubiquitin tag for sperm mitochondria. Nature. 1999;402:371–372. - PubMed
-
- Miller D. RNA in the ejaculate spermatozoon: a window into molecular events in spermatogenesis and a record of the unusual requirements of haploid gene expression and post-meiotic equilibration. Mol. Hum. Reprod. 1997;3:669–676. - PubMed
-
- Miller D, Ostermeier GC. Towards a better understanding of RNA carriage by ejaculate spermatozoa. Hum. Reprod. Update. 2006;12:757–767. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

