Evidence for a common mechanism of SIRT1 regulation by allosteric activators
- PMID: 23471411
- PMCID: PMC3799917
- DOI: 10.1126/science.1231097
Evidence for a common mechanism of SIRT1 regulation by allosteric activators
Abstract
A molecule that treats multiple age-related diseases would have a major impact on global health and economics. The SIRT1 deacetylase has drawn attention in this regard as a target for drug design. Yet controversy exists around the mechanism of sirtuin-activating compounds (STACs). We found that specific hydrophobic motifs found in SIRT1 substrates such as PGC-1α and FOXO3a facilitate SIRT1 activation by STACs. A single amino acid in SIRT1, Glu(230), located in a structured N-terminal domain, was critical for activation by all previously reported STAC scaffolds and a new class of chemically distinct activators. In primary cells reconstituted with activation-defective SIRT1, the metabolic effects of STACs were blocked. Thus, SIRT1 can be directly activated through an allosteric mechanism common to chemically diverse STACs.
Figures
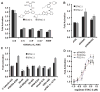
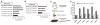


Comment in
-
Biochemistry. Red wine, toast of the town (again).Science. 2013 Mar 8;339(6124):1156-7. doi: 10.1126/science.1236463. Science. 2013. PMID: 23471392 Free PMC article. No abstract available.
-
Rejuvenating SIRT1 activators.Cell Metab. 2013 May 7;17(5):635-7. doi: 10.1016/j.cmet.2013.04.016. Cell Metab. 2013. PMID: 23663735
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

