A dynamic bronchial airway gene expression signature of chronic obstructive pulmonary disease and lung function impairment
- PMID: 23471465
- PMCID: PMC3707363
- DOI: 10.1164/rccm.201208-1449OC
A dynamic bronchial airway gene expression signature of chronic obstructive pulmonary disease and lung function impairment
Abstract
Rationale: Molecular phenotyping of chronic obstructive pulmonary disease (COPD) has been impeded in part by the difficulty in obtaining lung tissue samples from individuals with impaired lung function.
Objectives: We sought to determine whether COPD-associated processes are reflected in gene expression profiles of bronchial airway epithelial cells obtained by bronchoscopy.
Methods: Gene expression profiling of bronchial brushings obtained from 238 current and former smokers with and without COPD was performed using Affymetrix Human Gene 1.0 ST Arrays.
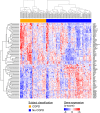
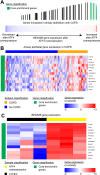
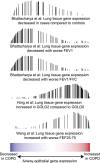
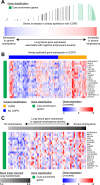
Measurements and main results: We identified 98 genes whose expression levels were associated with COPD status, FEV1% predicted, and FEV1/FVC. In silico analysis identified activating transcription factor 4 (ATF4) as a potential transcriptional regulator of genes with COPD-associated airway expression, and ATF4 overexpression in airway epithelial cells in vitro recapitulates COPD-associated gene expression changes. Genes with COPD-associated expression in the bronchial airway epithelium had similarly altered expression profiles in prior studies performed on small-airway epithelium and lung parenchyma, suggesting that transcriptomic alterations in the bronchial airway epithelium reflect molecular events found at more distal sites of disease activity. Many of the airway COPD-associated gene expression changes revert toward baseline after therapy with the inhaled corticosteroid fluticasone in independent cohorts.
Conclusions: Our findings demonstrate a molecular field of injury throughout the bronchial airway of active and former smokers with COPD that may be driven in part by ATF4 and is modifiable with therapy. Bronchial airway epithelium may ultimately serve as a relatively accessible tissue in which to measure biomarkers of disease activity for guiding clinical management of COPD.
Figures
Comment in
-
Chronic obstructive pulmonary disease genomics: yesterday, discovering population biomarkers; tomorrow, defining disease clusters.Am J Respir Crit Care Med. 2013 May 1;187(9):900-2. doi: 10.1164/rccm.201302-0340ED. Am J Respir Crit Care Med. 2013. PMID: 23634854 No abstract available.
References
-
- National Heart Lung and Blood Institute. NHLBI Fiscal Year 2010 Fact Book [2010; accessed 2013 Apr 10]. Available from: http://www.nhlbi.nih.gov/about/factbook-10/FactBook_2010.pdf.
-
- Murphy SL, Xu J, Kochanek KD. Deaths: preliminary data for 2012. National Vital Statistics Reports. Vol 60, No 4. Hyattsville, MD: National Center for Health Statistics; 2012.
-
- Spira A, Beane J, Pinto-Plata V, Kadar A, Liu G, Shah V, Celli B, Brody JS. Gene expression profiling of human lung tissue from smokers with severe emphysema. Am J Respir Cell Mol Biol. 2004;31:601–610. - PubMed
-
- Golpon HA, Coldren CD, Zamora MR, Cosgrove GP, Moore MD, Tuder RM, Geraci MW, Voelkel NF. Emphysema lung tissue gene expression profiling. Am J Respir Cell Mol Biol. 2004;31:595–600. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

