Cyclic di-GMP: the first 25 years of a universal bacterial second messenger
- PMID: 23471616
- PMCID: PMC3591986
- DOI: 10.1128/MMBR.00043-12
Cyclic di-GMP: the first 25 years of a universal bacterial second messenger
Abstract
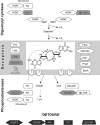
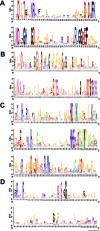
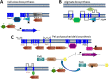
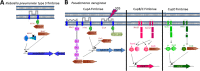
Twenty-five years have passed since the discovery of cyclic dimeric (3'→5') GMP (cyclic di-GMP or c-di-GMP). From the relative obscurity of an allosteric activator of a bacterial cellulose synthase, c-di-GMP has emerged as one of the most common and important bacterial second messengers. Cyclic di-GMP has been shown to regulate biofilm formation, motility, virulence, the cell cycle, differentiation, and other processes. Most c-di-GMP-dependent signaling pathways control the ability of bacteria to interact with abiotic surfaces or with other bacterial and eukaryotic cells. Cyclic di-GMP plays key roles in lifestyle changes of many bacteria, including transition from the motile to the sessile state, which aids in the establishment of multicellular biofilm communities, and from the virulent state in acute infections to the less virulent but more resilient state characteristic of chronic infectious diseases. From a practical standpoint, modulating c-di-GMP signaling pathways in bacteria could represent a new way of controlling formation and dispersal of biofilms in medical and industrial settings. Cyclic di-GMP participates in interkingdom signaling. It is recognized by mammalian immune systems as a uniquely bacterial molecule and therefore is considered a promising vaccine adjuvant. The purpose of this review is not to overview the whole body of data in the burgeoning field of c-di-GMP-dependent signaling. Instead, we provide a historic perspective on the development of the field, emphasize common trends, and illustrate them with the best available examples. We also identify unresolved questions and highlight new directions in c-di-GMP research that will give us a deeper understanding of this truly universal bacterial second messenger.
Figures




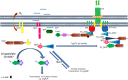


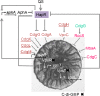
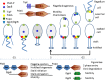

References
-
- Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279–281 - PubMed
-
- D'Argenio DA, Miller SI. 2004. Cyclic di-GMP as a bacterial second messenger. Microbiology 150:2497–2502 - PubMed
-
- Jenal U. 2004. Cyclic di-guanosine-monophosphate comes of age: a novel secondary messenger involved in modulating cell surface structures in bacteria? Curr. Opin. Microbiol. 7:185–191 - PubMed
-
- Jenal U, Malone J. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40:385–407 - PubMed
-
- Römling U, Gomelsky M, Galperin MY. 2005. c-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol. 57:629–639 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

