Diverse functions of restriction-modification systems in addition to cellular defense
- PMID: 23471617
- PMCID: PMC3591985
- DOI: 10.1128/MMBR.00044-12
Diverse functions of restriction-modification systems in addition to cellular defense
Abstract
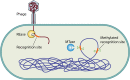
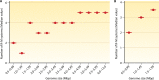
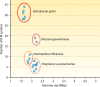
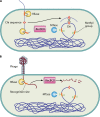
Restriction-modification (R-M) systems are ubiquitous and are often considered primitive immune systems in bacteria. Their diversity and prevalence across the prokaryotic kingdom are an indication of their success as a defense mechanism against invading genomes. However, their cellular defense function does not adequately explain the basis for their immaculate specificity in sequence recognition and nonuniform distribution, ranging from none to too many, in diverse species. The present review deals with new developments which provide insights into the roles of these enzymes in other aspects of cellular function. In this review, emphasis is placed on novel hypotheses and various findings that have not yet been dealt with in a critical review. Emerging studies indicate their role in various cellular processes other than host defense, virulence, and even controlling the rate of evolution of the organism. We also discuss how R-M systems could have successfully evolved and be involved in additional cellular portfolios, thereby increasing the relative fitness of their hosts in the population.
Figures


References
-
- Dryden DTF. 1999. Bacterial DNA methyltransferases, p 283–340 In Cheng X, Blumenthal RM. (ed), S-Adenosylmethionine-dependent methyltransferases: structures and functions. World Scientific Publishing, Singapore
-
- Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev SK, Dryden DTF, Dybvig K, Firman K, Gromova ES, Gumport RI, Halford SE, Hattman S, Heitman J, Hornby DP, Janulaitis A, Jeltsch A, Josephsen J, Kiss A, Klaenhammer TR, Kobayashi I, Kong H, Krüger DH, Lacks S, Marinus MG, Miyahara M, Morgan RD, Murray NE, Nagaraja V, Piekarowicz A, Pingoud A, Raleigh E, Rao DN, Reich N, Repin VE, Selker EU, Shaw PC, Stein DC, Stoddard BL, Szybalski W, Trautner TA, Van Etten JL, Vitor JM, Wilson GG, Xu SY. 2003. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 31: 1805–1812 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

