A polymetamorphic protein
- PMID: 23471712
- PMCID: PMC3649265
- DOI: 10.1002/pro.2248
A polymetamorphic protein
Abstract
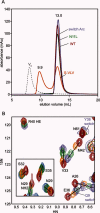
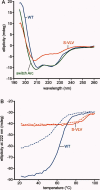
Arc repressor is a homodimeric protein with a ribbon-helix-helix fold. A single polar-to-hydrophobic substitution (N11L) at a solvent-exposed position leads to population of an alternate dimeric fold in which 3₁₀ helices replace a β-sheet. Here we find that the variant Q9V/N11L/R13V (S-VLV), with two additional polar-to-hydrophobic surface mutations in the same β-sheet, forms a highly stable, reversibly folded octamer with approximately half the α-helical content of wild-type Arc. At low protein concentration and low ionic strength, S-VLV also populates both dimeric topologies previously observed for N11L, as judged by NMR chemical shift comparisons. Thus, accumulation of simple hydrophobic mutations in Arc progressively reduces fold specificity, leading first to a sequence with two folds and then to a manifold bridge sequence with at least three different topologies. Residues 9-14 of S-VLV form a highly hydrophobic stretch that is predicted to be amyloidogenic, but we do not observe aggregates of higher order than octamer. Increases in sequence hydrophobicity can promote amyloid aggregation but also exert broader and more complex effects on fold specificity. Altered native folds, changes in fold coupled to oligomerization, toxic pre-amyloid oligomers, and amyloid fibrils may represent a near continuum of accessible alternatives in protein structure space.
Copyright © 2013 The Protein Society.
Figures



References
-
- Chan HS, Bornberg-Bauer E. Perspectives on protein evolution from simple exact models. Appl Bioinformatics. 2002;1:121–144. - PubMed
-
- Murzin AG. Metamorphic proteins. Science. 2008;320:1725–1726. - PubMed
-
- Cordes MHJ, Stewart KL. The porous borders of the protein world. Structure. 2012;20:199–200. - PubMed
-
- Meier S, Ozbek S. A biological cosmos of parallel universes: does protein structural plasticity facilitate evolution? Bioessays. 2007;29:1095–1104. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

