Lysophosphatidylinositol-acyltransferase-1 (LPIAT1) is required to maintain physiological levels of PtdIns and PtdInsP(2) in the mouse
- PMID: 23472195
- PMCID: PMC3589398
- DOI: 10.1371/journal.pone.0058425
Lysophosphatidylinositol-acyltransferase-1 (LPIAT1) is required to maintain physiological levels of PtdIns and PtdInsP(2) in the mouse
Abstract
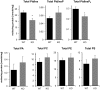
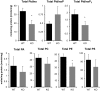
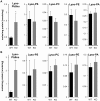
We disrupted the gene encoding lysophosphatidylinositol-acyltransferase-1 (LPIAT1) in the mouse with the aim of understanding its role in determining cellular phosphoinositide content. LPIAT1(-/-) mice were born at lower than Mendelian ratios and exhibited a severe developmental brain defect. We compared the phospholipid content of livers and brains from LPIAT1(-/-) and LPIAT1(+/+) littermates by LC-ESI/MS. In accord with previous studies, the most abundant molecular species of each phosphoinositide class (PtdIns, PtdInsP, PtdInsP2 and PtdInsP3) possessed a C38∶4 complement of fatty-acyl esters (C18∶0 and C20∶4 are usually assigned to the sn-1 and sn-2 positions, respectively). LPIAT1(-/-) liver and brain contained relatively less of the C38∶4 species of PtdIns, PtdInsP and PtdInsP2 (dropping from 95-97% to 75-85% of the total species measured for each lipid class) and relatively more of the less abundant species (PtdInsP3 less abundant species were below our quantification levels). The increases in the less abundant PtdIns and PtdInsP2 species did not compensate for the loss in C38∶4 species, resulting in a 26-44% reduction in total PtdIns and PtdInsP2 levels in both brain and liver. LPIAT1(-/-) brain and liver also contained increased levels of C18∶0 lyso-PtdIns (300% and 525% respectively) indicating a defect in the reacylation of this molecule. LPIAT1(-/-) brain additionally contained significantly reduced C38∶4 PC and PE levels (by 47% and 55% respectively), possibly contributing to the phenotype in this organ. The levels of all other molecular species of PC, PE, PS and PA measured in the brain and liver were very similar between LPIAT1(-/-) and LPIAT1(+/+) samples. These results suggest LPIAT1 activity plays a non-redundant role in maintaining physiological levels of PtdIns within an active deacylation/reacylation cycle in mouse tissues. They also suggest that this pathway must act in concert with other, as yet unidentified, mechanisms to achieve the enrichment observed in C38∶4 molecular species of phosphoinositides.
Conflict of interest statement
Figures





References
-
- Kudo I, Murakami M (2002) Phospholipase A2 enzymes. Prostaglandins Other Lipid Mediat 68–69: 3–58. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- WT_/Wellcome Trust/United Kingdom
- BBS/E/B/000C0413/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/B/000C0415/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J004456/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

