Epigenetic hierarchy within the MAGEA1 cancer-germline gene: promoter DNA methylation dictates local histone modifications
- PMID: 23472218
- PMCID: PMC3589373
- DOI: 10.1371/journal.pone.0058743
Epigenetic hierarchy within the MAGEA1 cancer-germline gene: promoter DNA methylation dictates local histone modifications
Abstract
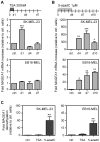
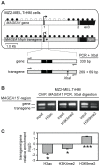
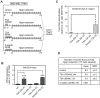
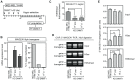
Gene MAGEA1 belongs to a group of human germline-specific genes that rely on DNA methylation for repression in somatic tissues. Many of these genes, termed cancer-germline (CG) genes, become demethylated and activated in a wide variety of tumors, where they encode tumor-specific antigens. The process leading to DNA demethylation of CG genes in tumors remains unclear. Previous data suggested that histone acetylation might be involved. Here, we investigated the relative contribution of DNA methylation and histone acetylation in the epigenetic regulation of gene MAGEA1. We show that MAGEA1 DNA hypomethylation in expressing melanoma cells is indeed correlated with local increases in histone H3 acetylation (H3ac). However, when MAGEA1-negative cells were exposed to a histone deacetylase inhibitor (TSA), we observed only short-term activation of the gene and detected no demethylation of its promoter. As a more sensitive assay, we used a cell clone harboring a methylated MAGEA1/hph construct, which confers resistance to hygromycin upon stable re-activation. TSA induced only transient de-repression of the transgene, and did not lead to the emergence of hygromycin-resistant cells. In striking contrast, transient depletion of DNA-methyltransferase-1 in the reporter cell clone gave rise to a hygromycin-resistant population, in which the re-activated MAGEA1/hph transgene displayed not only marked DNA hypomethylation, but also significant reversal of histone marks, including gains in H3ac and H3K4me2, and losses of H3K9me2. Collectively, our results indicate that DNA methylation has a dominant role in the epigenetic hierarchy governing MAGEA1 expression.
Conflict of interest statement
Figures

References
-
- Bird A (2002) DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21. - PubMed
-
- Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP (1998) Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res 72: 141–196. - PubMed
-
- Esteller M (2008) Epigenetics in cancer. N Engl J Med 358: 1148–1159. - PubMed
-
- Ehrlich M (2002) DNA methylation in cancer: too much, but also too little. Oncogene 21: 5400–5413. - PubMed
-
- Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, et al. (2003) Induction of tumors in mice by genomic hypomethylation. Science 300: 489–492. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

