The hierarchy of the 3D genome
- PMID: 23473598
- PMCID: PMC3741673
- DOI: 10.1016/j.molcel.2013.02.011
The hierarchy of the 3D genome
Abstract
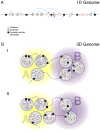
Mammalian genomes encode genetic information in their linear sequence, but appropriate expression of their genes requires chromosomes to fold into complex three-dimensional structures. Transcriptional control involves the establishment of physical connections among genes and regulatory elements, both along and between chromosomes. Recent technological innovations in probing the folding of chromosomes are providing new insights into the spatial organization of genomes and its role in gene regulation. It is emerging that folding of large complex chromosomes involves a hierarchy of structures, from chromatin loops that connect genes and enhancers to larger chromosomal domains and nuclear compartments. The larger these structures are along this hierarchy, the more stable they are within cells, while becoming more stochastic between cells. Here, we review the experimental and theoretical data on this hierarchy of structures and propose a key role for the recently discovered topologically associating domains.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures


References
-
- Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, Tixier V, Mas A, Cavalli G. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell. 2011;144:214–226. - PubMed
-
- Bau D, Marti-Renom MA. Structure determination of genomic domains by satisfaction of spatial restraints. Chromosome research: an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2011;19:25–35. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

