Increased genome instability in human DNA segments with self-chains: homology-induced structural variations via replicative mechanisms
- PMID: 23474816
- PMCID: PMC3674805
- DOI: 10.1093/hmg/ddt113
Increased genome instability in human DNA segments with self-chains: homology-induced structural variations via replicative mechanisms
Abstract
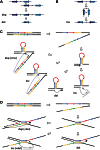
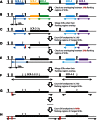
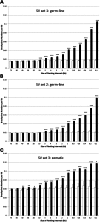
Environmental factors including ionizing radiation and chemical agents have been known to be able to induce DNA rearrangements and cause genomic structural variations (SVs); however, the roles of intrinsic characteristics of the human genome, such as regional genome architecture, in SV formation and the potential mechanisms underlying genomic instability remain to be further elucidated. Recently, locus-specific observations showed that 'self-chain' (SC), a group of short low-copy repeats (LCRs) in the human genome, can induce autism-associated SV mutations of the MECP2 and NRXN1 genes. In this study, we conducted a genome-wide analysis to investigate SCs and their potential roles in genomic SV formation. Utilizing a vast amount of human SV data, we observed a significant biased distribution of human germline SV breakpoints to SC regions. Notably, the breakpoint distribution pattern is different between SV types across deletion, duplication, inversion and insertion. Our observations were coincident with a mechanism of SC-induced DNA replicative errors, whereas SC may sporadically be used as substrates of nonallelic homologous recombination (NAHR). This contention was further supported by our consistent findings in somatic SV mutations of cancer genomes, suggesting a general mechanism of SC-induced genome instability in human germ and somatic cells.
Figures


References
-
- Stankiewicz P., Lupski J.R. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 2010;61:437–455. - PubMed
-
- Alkan C., Coe B.P., Eichler E.E. Genome structural variation discovery and genotyping. Nat. Rev. Genet. 2011;12:363–376. doi:10.1080/13803390701336411. - DOI - PMC - PubMed
-
- The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi:10.1080/13803390701346113. - DOI - PMC - PubMed
-
- Lupski J.R. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi:10.1002/(SICI)1099-1166(199706)12:6<619::AID-GPS554>3.0.CO;2-H. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

