Oct4 switches partnering from Sox2 to Sox17 to reinterpret the enhancer code and specify endoderm
- PMID: 23474895
- PMCID: PMC3616284
- DOI: 10.1038/emboj.2013.31
Oct4 switches partnering from Sox2 to Sox17 to reinterpret the enhancer code and specify endoderm
Abstract
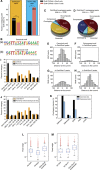
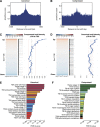
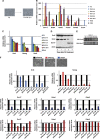
How regulatory information is encoded in the genome is poorly understood and poses a challenge when studying biological processes. We demonstrate here that genomic redistribution of Oct4 by alternative partnering with Sox2 and Sox17 is a fundamental regulatory event of endodermal specification. We show that Sox17 partners with Oct4 and binds to a unique 'compressed' Sox/Oct motif that earmarks endodermal genes. This is in contrast to the pluripotent state where Oct4 selectively partners with Sox2 at 'canonical' binding sites. The distinct selection of binding sites by alternative Sox/Oct partnering is underscored by our demonstration that rationally point-mutated Sox17 partners with Oct4 on pluripotency genes earmarked by the canonical Sox/Oct motif. In an endodermal differentiation assay, we demonstrate that the compressed motif is required for proper expression of endodermal genes. Evidently, Oct4 drives alternative developmental programs by switching Sox partners that affects enhancer selection, leading to either an endodermal or pluripotent cell fate. This work provides insights in understanding cell fate transcriptional regulation by highlighting the direct link between the DNA sequence of an enhancer and a developmental outcome.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





Comment in
-
Oct4 shuffles Sox partners to direct cell fate.EMBO J. 2013 Apr 3;32(7):917-9. doi: 10.1038/emboj.2013.48. Epub 2013 Mar 8. EMBO J. 2013. PMID: 23474893 Free PMC article.
References
-
- Barbacci E, Reber M, Ott MO, Breillat C, Huetz F, Cereghini S (1999) Variant hepatocyte nuclear factor 1 is required for visceral endoderm specification. Development 126: 4795–4805 - PubMed
-
- Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R (2006) Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis 44: 23–28 - PubMed
-
- Biggin MD (2011) Animal transcription networks as highly connected, quantitative continua. Dev Cell 21: 611–626 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

