A scale-space method for detecting recurrent DNA copy number changes with analytical false discovery rate control
- PMID: 23476020
- PMCID: PMC3643574
- DOI: 10.1093/nar/gkt155
A scale-space method for detecting recurrent DNA copy number changes with analytical false discovery rate control
Abstract
Tumor formation is partially driven by DNA copy number changes, which are typically measured using array comparative genomic hybridization, SNP arrays and DNA sequencing platforms. Many techniques are available for detecting recurring aberrations across multiple tumor samples, including CMAR, STAC, GISTIC and KC-SMART. GISTIC is widely used and detects both broad and focal (potentially overlapping) recurring events. However, GISTIC performs false discovery rate control on probes instead of events. Here we propose Analytical Multi-scale Identification of Recurrent Events, a multi-scale Gaussian smoothing approach, for the detection of both broad and focal (potentially overlapping) recurring copy number alterations. Importantly, false discovery rate control is performed analytically (no need for permutations) on events rather than probes. The method does not require segmentation or calling on the input dataset and therefore reduces the potential loss of information due to discretization. An important characteristic of the approach is that the error rate is controlled across all scales and that the algorithm outputs a single profile of significant events selected from the appropriate scales. We perform extensive simulations and showcase its utility on a glioblastoma SNP array dataset. Importantly, ADMIRE detects focal events that are missed by GISTIC, including two events involving known glioma tumor-suppressor genes: CDKN2C and NF1.
Figures

 and the auto-correlation r. Panel C shows the kernel convolution per scale. In this illustration, we propose to repeat the steps in Panels A, B and C one thousand times to obtain an empirical approximation of the null distribution and use these distributions to derive a threshold per scale corresponding to the desired control of FDR and FWER. However, in this article, we derive an analytical relationship between the thresholds and FWER or FDR.
and the auto-correlation r. Panel C shows the kernel convolution per scale. In this illustration, we propose to repeat the steps in Panels A, B and C one thousand times to obtain an empirical approximation of the null distribution and use these distributions to derive a threshold per scale corresponding to the desired control of FDR and FWER. However, in this article, we derive an analytical relationship between the thresholds and FWER or FDR.
 found across the whole genome (as predicted by the null hypothesis). The threshold is selected at
found across the whole genome (as predicted by the null hypothesis). The threshold is selected at  , a close upper-bound for the FWER of 0.01.
, a close upper-bound for the FWER of 0.01.
 and r) is restricted to
and r) is restricted to  , as illustrated by the dotted line at the top of the figure. (B) On recursive level 2, we follow the exact same procedure, except this time, estimate the null parameters in the broad event
, as illustrated by the dotted line at the top of the figure. (B) On recursive level 2, we follow the exact same procedure, except this time, estimate the null parameters in the broad event  . This allows us to detect embedded focal events inside broader events.
. This allows us to detect embedded focal events inside broader events.
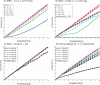
 (x-axis) and that measured across 1000 simulations (y-axis) of aCGH profiles containing only passenger events. (A) We fix the kernel width to be small (40 kb) and the SNR at 1 to represent measurement noise. We vary the number of samples to aggregate for each simulation experiment. (B) A similar experiment on simulated aCGH profiles where we added no measurement noise (
(x-axis) and that measured across 1000 simulations (y-axis) of aCGH profiles containing only passenger events. (A) We fix the kernel width to be small (40 kb) and the SNR at 1 to represent measurement noise. We vary the number of samples to aggregate for each simulation experiment. (B) A similar experiment on simulated aCGH profiles where we added no measurement noise ( ) and therefore effectively work with segmented samples. The black line depicts the result obtained when using cyclic permutation to create a null hypothesis on the glioma dataset. (C) The number of simulated samples to aggregate is fixed at 100 and the kernel width is varied, showing good theoretical predictions for all kernels. The black line indicates the mean number of events detected when we apply multi-scale selection. (D) Similar results are depicted when using cyclic permutations to create a null hypothesis on the glioma dataset. The genome size for the simulated data is only
) and therefore effectively work with segmented samples. The black line depicts the result obtained when using cyclic permutation to create a null hypothesis on the glioma dataset. (C) The number of simulated samples to aggregate is fixed at 100 and the kernel width is varied, showing good theoretical predictions for all kernels. The black line indicates the mean number of events detected when we apply multi-scale selection. (D) Similar results are depicted when using cyclic permutations to create a null hypothesis on the glioma dataset. The genome size for the simulated data is only  bps, whereas the glioma dataset consists of all probes stretching from chromosome 1 to 22. Error bars indicate the standard error of the empirical
bps, whereas the glioma dataset consists of all probes stretching from chromosome 1 to 22. Error bars indicate the standard error of the empirical  .
.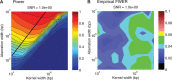
 at 5%. The black line indicates the maximum allowed kernel width at which an aberration can be detected if we apply filtering with
at 5%. The black line indicates the maximum allowed kernel width at which an aberration can be detected if we apply filtering with  in the multi-scale methodology. See
in the multi-scale methodology. See 
 , while keeping the number of samples to aggregate per simulation fixed at 200, i.e.
, while keeping the number of samples to aggregate per simulation fixed at 200, i.e.  . Furthermore, we do not add any noise, as the
. Furthermore, we do not add any noise, as the  , implying that all samples are segmented. (B) The empirical FDR (left panel) and power (right panel) as a function of the number of samples to aggregate S for the SNR assuming the following values,
, implying that all samples are segmented. (B) The empirical FDR (left panel) and power (right panel) as a function of the number of samples to aggregate S for the SNR assuming the following values,  , while keeping the number of focal recurrent events and FDR fixed at 50 (
, while keeping the number of focal recurrent events and FDR fixed at 50 ( ) and 5%, respectively.
) and 5%, respectively.
References
-
- Rouveirol C, Stransky N, Hupé P, La Rosa P, Viara E, Barillot E, Radvanyi F. Computation of recurrent minimal genomic alterations from array-CGH data. Bioinformatics. 2006;22:849–856. - PubMed
-
- Shah SP, Lam WL, Ng RT, Murphy KP. Modeling recurrent DNA copy number alterations in array CGH data. Bioinformatics. 2007;23:i450–i458. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

