Homologous recombination DNA repair genes play a critical role in reprogramming to a pluripotent state
- PMID: 23478019
- PMCID: PMC4315363
- DOI: 10.1016/j.celrep.2013.02.005
Homologous recombination DNA repair genes play a critical role in reprogramming to a pluripotent state
Abstract
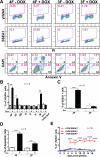
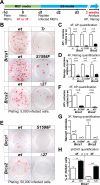
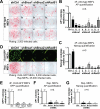
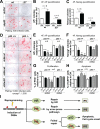
Induced pluripotent stem cells (iPSCs) hold great promise for personalized regenerative medicine. However, recent studies show that iPSC lines carry genetic abnormalities, suggesting that reprogramming may be mutagenic. Here, we show that the ectopic expression of reprogramming factors increases the level of phosphorylated histone H2AX, one of the earliest cellular responses to DNA double-strand breaks (DSBs). Additional mechanistic studies uncover a direct role of the homologous recombination (HR) pathway, a pathway essential for error-free repair of DNA DSBs, in reprogramming. This role is independent of the use of integrative or nonintegrative methods in introducing reprogramming factors, despite the latter being considered a safer approach that circumvents genetic modifications. Finally, deletion of the tumor suppressor p53 rescues the reprogramming phenotype in HR-deficient cells primarily through the restoration of reprogramming-dependent defects in cell proliferation and apoptosis. These mechanistic insights have important implications for the design of safer approaches to creating iPSCs.
Copyright © 2013 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
References
-
- Daley GQ. Common themes of dedifferentiation in somatic cell reprogramming and cancer. Cold Spring Harbor symposia on quantitative biology. 2008;73:171–174. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

