Protection of neuronal diversity at the expense of neuronal numbers during nutrient restriction in the Drosophila visual system
- PMID: 23478023
- PMCID: PMC3617362
- DOI: 10.1016/j.celrep.2013.02.006
Protection of neuronal diversity at the expense of neuronal numbers during nutrient restriction in the Drosophila visual system
Abstract
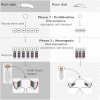
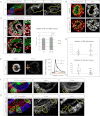
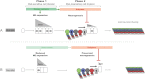
Systemic signals provided by nutrients and hormones are known to coordinate the growth and proliferation of different organs during development. However, within the brain, it is unclear how these signals influence neural progenitor divisions and neuronal diversity. Here, in the Drosophila visual system, we identify two developmental phases with different sensitivities to dietary nutrients. During early larval stages, nutrients regulate the size of the neural progenitor pool via insulin/PI3K/TOR-dependent symmetric neuroepithelial divisions. During late larval stages, neural proliferation becomes insensitive to dietary nutrients, and the steroid hormone ecdysone acts on Delta/Notch signaling to promote the switch from symmetric mitoses to asymmetric neurogenic divisions. This mechanism accounts for why sustained undernourishment during visual system development restricts neuronal numbers while protecting neuronal diversity. These studies reveal an adaptive mechanism that helps to retain a functional visual system over a range of different brain sizes in the face of suboptimal nutrition.
Copyright © 2013 The Authors. Published by Elsevier Inc. All rights reserved.
Figures






References
-
- Brennan C.A., Ashburner M., Moses K. Ecdysone pathway is required for furrow progression in the developing Drosophila eye. Development. 1998;125:2653–2664. - PubMed
-
- Britton J.S., Edgar B.A. Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development. 1998;125:2149–2158. - PubMed
-
- Brogiolo W., Stocker H., Ikeya T., Rintelen F., Fernandez R., Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 2001;11:213–221. - PubMed
-
- Brown H.L., Cherbas L., Cherbas P., Truman J.W. Use of time-lapse imaging and dominant negative receptors to dissect the steroid receptor control of neuronal remodeling in Drosophila. Development. 2006;133:275–285. - PubMed
Supplemental References
-
- Awad T.A., Truman J.W. Postembryonic development of the midline glia in the CNS of Drosophila: proliferation, programmed cell death, and endocrine regulation. Dev. Biol. 1997;187:283–297. - PubMed
-
- Bainbridge S.P., Bownes M. Staging the metamorphosis of Drosophila melanogaster. J. Embryol. Exp. Morphol. 1981;66:57–80. - PubMed
-
- Currie D.A., Milner M.J., Evans C.W. The growth and differentiation in vitro of leg and wing imaginal disc cells from Drosophila melanogaster. Development. 1988;102:805–814.
-
- Lee T., Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. - PubMed
-
- Neubueser D., Warren J.T., Gilbert L.I., Cohen S.M. Molting defective is required for ecdysone biosynthesis. Dev. Biol. 2005;280:362–372. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

