Human DEAD box helicase 3 couples IκB kinase ε to interferon regulatory factor 3 activation
- PMID: 23478265
- PMCID: PMC3647972
- DOI: 10.1128/MCB.01603-12
Human DEAD box helicase 3 couples IκB kinase ε to interferon regulatory factor 3 activation
Abstract
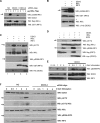
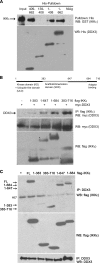
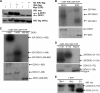
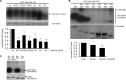
The human DEAD box protein 3 (DDX3) has been implicated in different processes contributing to gene expression. Interestingly, DDX3 is required as an essential host factor for the replication of HIV and hepatitis C virus (HCV) and is therefore considered a potential drug target. On the other hand, DDX3 interacts with IκB kinase ε (IKKε) and TANK-binding kinase 1 (TBK1) and contributes to the induction of antiviral type I interferons (IFNs). However, the molecular mechanism by which DDX3 contributes to IFN induction remains unclear. Here we show that DDX3 mediates phosphorylation of interferon regulatory factor 3 (IRF3) by the kinase IKKε. DDX3 directly interacts with IKKε and enhances its autophosphorylation and activation. IKKε then phosphorylates several serine residues in the N terminus of DDX3. Phosphorylation of DDX3 at serine 102 (S102) is required for recruitment of IRF3 to DDX3, facilitating its phosphorylation by IKKε. Mutation of S102 to alanine disrupted the interaction between DDX3 and IRF3 but not that between DDX3 and IKKε. The S102A mutation failed to enhance ifnb promoter activation, suggesting that the DDX3-IRF3 interaction is crucial for this effect. Our data implicates DDX3 as a scaffolding adaptor that directly facilitates phosphorylation of IRF3 by IKKε. DDX3 might thus be involved in pathway-specific activation of IRF3.
Figures


Similar articles
-
Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation.EMBO J. 2008 Aug 6;27(15):2147-57. doi: 10.1038/emboj.2008.143. Epub 2008 Jul 17. EMBO J. 2008. PMID: 18636090 Free PMC article.
-
DDX3 directly facilitates IKKα activation and regulates downstream signalling pathways.Biochem J. 2018 Nov 20;475(22):3595-3607. doi: 10.1042/BCJ20180163. Biochem J. 2018. PMID: 30341167
-
DDX3 directly regulates TRAF3 ubiquitination and acts as a scaffold to co-ordinate assembly of signalling complexes downstream from MAVS.Biochem J. 2017 Feb 15;474(4):571-587. doi: 10.1042/BCJ20160956. Epub 2016 Dec 15. Biochem J. 2017. PMID: 27980081
-
Regulation of IRF3 activation in human antiviral signaling pathways.Biochem Pharmacol. 2022 Jun;200:115026. doi: 10.1016/j.bcp.2022.115026. Epub 2022 Mar 31. Biochem Pharmacol. 2022. PMID: 35367198 Review.
-
RNA helicase DDX3: at the crossroad of viral replication and antiviral immunity.Rev Med Virol. 2015 Sep;25(5):286-99. doi: 10.1002/rmv.1845. Epub 2015 Jul 14. Rev Med Virol. 2015. PMID: 26174373 Review.
Cited by
-
RNA helicase DDX3 regulates RAD51 localization and DNA damage repair in Ewing sarcoma.iScience. 2024 Jan 16;27(2):108925. doi: 10.1016/j.isci.2024.108925. eCollection 2024 Feb 16. iScience. 2024. PMID: 38323009 Free PMC article.
-
The multifaceted roles of NLRP3-modulating proteins in virus infection.Front Immunol. 2022 Aug 30;13:987453. doi: 10.3389/fimmu.2022.987453. eCollection 2022. Front Immunol. 2022. PMID: 36110852 Free PMC article. Review.
-
Tyrosine phosphorylation of IRF3 by BLK facilitates its sufficient activation and innate antiviral response.PLoS Pathog. 2023 Oct 23;19(10):e1011742. doi: 10.1371/journal.ppat.1011742. eCollection 2023 Oct. PLoS Pathog. 2023. PMID: 37871014 Free PMC article.
-
DDX3 DEAD-box RNA helicase is a host factor that restricts hepatitis B virus replication at the transcriptional level.J Virol. 2014 Dec;88(23):13689-98. doi: 10.1128/JVI.02035-14. Epub 2014 Sep 17. J Virol. 2014. PMID: 25231298 Free PMC article.
-
CIKS/DDX3X interaction controls the stability of the Zc3h12a mRNA induced by IL-17.J Immunol. 2015 Apr 1;194(7):3286-94. doi: 10.4049/jimmunol.1401589. Epub 2015 Feb 20. J Immunol. 2015. PMID: 25710910 Free PMC article.
References
-
- Schröder M. 2010. Human DEAD-box protein 3 has multiple functions in gene regulation and cell cycle control and is a prime target for viral manipulation. Biochem. Pharmacol. 79:297–306 - PubMed
-
- Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD, Chien M, Weir DB, Russo JJ, Ju J, Brownstein MJ, Sheridan R, Sander C, Zavolan M, Tuschl T, Rice CM. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. U. S. A. 104:12884–12889 - PMC - PubMed
-
- Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT. 2004. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 119:381–392 - PubMed
-
- Oshiumi H, Sakai K, Matsumoto M, Seya T. 2010. DEAD/H BOX 3 (DDX3) helicase binds the RIG-I adaptor IPS-1 to up-regulate IFN-beta-inducing potential. Eur. J. Immunol. 40:940–948 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
