Genomic signatures of selection at linked sites: unifying the disparity among species
- PMID: 23478346
- PMCID: PMC4066956
- DOI: 10.1038/nrg3425
Genomic signatures of selection at linked sites: unifying the disparity among species
Abstract
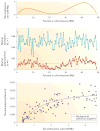
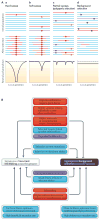
Population genetics theory supplies powerful predictions about how natural selection interacts with genetic linkage to sculpt the genomic landscape of nucleotide polymorphism. Both the spread of beneficial mutations and the removal of deleterious mutations act to depress polymorphism levels, especially in low-recombination regions. However, empiricists have documented extreme disparities among species. Here we characterize the dominant features that could drive differences in linked selection among species--including roles for selective sweeps being 'hard' or 'soft'--and the concealing effects of demography and confounding genomic variables. We advocate targeted studies of closely related species to unify our understanding of how selection and linkage interact to shape genome evolution.
Figures

References
-
- Wiehe THE, Stephan W. Analysis of a genetic hitchhiking model, and its application to DNA polymorphism data from Drosophila melanogaster. Mol Biol Evol. 1993;10:842–854. - PubMed
-
- Maynard Smith J, Haigh J. Hitch-hiking effect of a favorable gene. Genet Res. 1974;23:23–35. The classic theoretical study describing the process of genetic hitchhiking. - PubMed
-
- Hill WG, Robertson A. Effect of linkage on limits to artificial selection. Genet Res. 1966;8:269–294. The classic model of how linked selected loci interfere with each other’s ability to fix the favored alleles in the population. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

