Understanding bacteriophage specificity in natural microbial communities
- PMID: 23478639
- PMCID: PMC3705297
- DOI: 10.3390/v5030806
Understanding bacteriophage specificity in natural microbial communities
Abstract
Studying the coevolutionary dynamics between bacteria and the bacteriophage viruses that infect them is critical to understanding both microbial diversity and ecosystem functioning. Phages can play a key role in shaping bacterial population dynamics and can significantly alter both intra- and inter-specific competition among bacterial hosts. Predicting how phages might influence community stability and apparent competition, however, requires an understanding of how bacteria-phage interaction networks evolve as a function of host diversity and community dynamics. Here, we first review the progress that has been made in understanding phage specificity, including the use of experimental evolution, we then introduce a new dataset on natural bacteriophages collected from the phyllosphere of horse chestnut trees, and finally we highlight that bacterial sensitivity to phage is rarely a binary trait and that this variation should be taken into account and reported. We emphasize that there is currently insufficient evidence to make broad generalizations about phage host range in natural populations, the limits of phage adaptation to novel hosts, or the implications of phage specificity in shaping microbial communities. However, the combination of experimental and genomic approaches with the study of natural communities will allow new insight to the evolution and impact of phage specificity within complex bacterial communities.
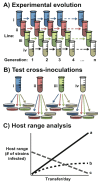
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

