Single-cell gene expression profiling reveals functional heterogeneity of undifferentiated human epidermal cells
- PMID: 23482486
- PMCID: PMC3596987
- DOI: 10.1242/dev.087551
Single-cell gene expression profiling reveals functional heterogeneity of undifferentiated human epidermal cells
Abstract
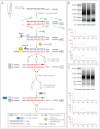
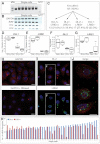
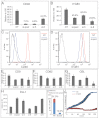
Human epidermal stem cells express high levels of β1 integrins, delta-like 1 (DLL1) and the EGFR antagonist LRIG1. However, there is cell-to-cell variation in the relative abundance of DLL1 and LRIG1 mRNA transcripts. Single-cell global gene expression profiling showed that undifferentiated cells fell into two clusters delineated by expression of DLL1 and its binding partner syntenin. The DLL1(+) cluster had elevated expression of genes associated with endocytosis, integrin-mediated adhesion and receptor tyrosine kinase signalling. Differentially expressed genes were not independently regulated, as overexpression of DLL1 alone or together with LRIG1 led to the upregulation of other genes in the DLL1(+) cluster. Overexpression of DLL1 and LRIG1 resulted in enhanced extracellular matrix adhesion and increased caveolin-dependent EGFR endocytosis. Further characterisation of CD46, one of the genes upregulated in the DLL1(+) cluster, revealed it to be a novel cell surface marker of human epidermal stem cells. Cells with high endogenous levels of CD46 expressed high levels of β1 integrin and DLL1 and were highly adhesive and clonogenic. Knockdown of CD46 decreased proliferative potential and β1 integrin-mediated adhesion. Thus, the previously unknown heterogeneity revealed by our studies results in differences in the interaction of undifferentiated basal keratinocytes with their environment.
Figures





References
-
- Adams J. C., Watt F. M. (1989). Fibronectin inhibits the terminal differentiation of human keratinocytes. Nature 340, 307–309 - PubMed
-
- Bauwens C. L., Song H., Thavandiran N., Ungrin M., Massé S., Nanthakumar K., Seguin C., Zandstra P. W. (2011). Geometric control of cardiomyogenic induction in human pluripotent stem cells. Tissue Eng. Part A 17, 1901–1909 - PubMed
-
- Beekman J. M., Coffer P. J. (2008). The ins and outs of syntenin, a multifunctional intracellular adaptor protein. J. Cell Sci. 121, 1349–1355 - PubMed
-
- Campos L. S., Decker L., Taylor V., Skarnes W. (2006). Notch, epidermal growth factor receptor, and beta1-integrin pathways are coordinated in neural stem cells. J. Biol. Chem. 281, 5300–5309 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

