Patterns of regulatory activity across diverse human cell types predict tissue identity, transcription factor binding, and long-range interactions
- PMID: 23482648
- PMCID: PMC3638134
- DOI: 10.1101/gr.152140.112
Patterns of regulatory activity across diverse human cell types predict tissue identity, transcription factor binding, and long-range interactions
Abstract
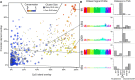
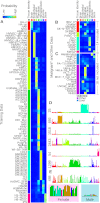
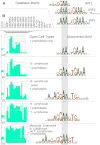
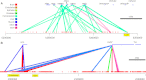
Regulatory elements recruit transcription factors that modulate gene expression distinctly across cell types, but the relationships among these remains elusive. To address this, we analyzed matched DNase-seq and gene expression data for 112 human samples representing 72 cell types. We first defined more than 1800 clusters of DNase I hypersensitive sites (DHSs) with similar tissue specificity of DNase-seq signal patterns. We then used these to uncover distinct associations between DHSs and promoters, CpG islands, conserved elements, and transcription factor motif enrichment. Motif analysis within clusters identified known and novel motifs in cell-type-specific and ubiquitous regulatory elements and supports a role for AP-1 regulating open chromatin. We developed a classifier that accurately predicts cell-type lineage based on only 43 DHSs and evaluated the tissue of origin for cancer cell types. A similar classifier identified three sex-specific loci on the X chromosome, including the XIST lincRNA locus. By correlating DNase I signal and gene expression, we predicted regulated genes for more than 500K DHSs. Finally, we introduce a web resource to enable researchers to use these results to explore these regulatory patterns and better understand how expression is modulated within and across human cell types.
Figures



References
-
- Angel P, Hess J 2012. The multi-gene family of transcription factor AP-1. In Regulation of organelle and cell compartment signaling: Cell signaling collection (ed. Bradshaw RA, Dennis EA), pp. 53–62. Academic Press, San Diego
-
- Angel P, Karin M 1991. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta 1072: 129–157 - PubMed
-
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
