Orai1-NFAT signalling pathway triggered by T cell receptor stimulation
- PMID: 23483280
- PMCID: PMC3887911
- DOI: 10.1007/s10059-013-0073-2
Orai1-NFAT signalling pathway triggered by T cell receptor stimulation
Abstract
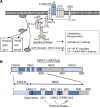
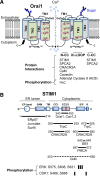
T cell receptor (TCR) stimulation plays a crucial role in development, homeostasis, proliferation, cell death, cytokine production, and differentiation of T cells. Thus, in depth understanding of TCR signalling is crucial for development of therapy targeting inflammatory diseases, improvement of vaccination efficiency, and cancer therapy utilizing T cell-based strategies. TCR activation turns on various signalling pathways, one of the important one being the Ca(2+)-calcineurin-nuclear factor of activated T cells (NFAT) signalling pathway. Stimulation of TCRs triggers depletion of intracellular Ca(2+) store and in turn, initiates store-operated Ca(2+) entry (SOCE), one of the major mechanisms to raise the intracellular Ca(2+) concentrations in T cells. Ca(2+)-release-activated-Ca(2+) (CRAC) channels are a prototype of store-operated Ca(2+) (SOC) channels in immune cells that are very well characterized. Recent identification of STIM1 as the endoplasmic reticulum (ER) Ca(2+) sensor and Orai1 as the pore subunit has dramatically advanced the understanding of CRAC channels and provides a molecular tool to investigate the physiological outcomes of Ca(2+) signalling during immune responses. In this review, we focus on our current understanding of CRAC channel activation, regulation, and downstream calcineurin-NFAT signaling pathway.
Figures



References
-
- Aramburu J, Yaffe MB, Lopez-Rodriguez C, Cantley LC, Hogan PG, Rao A. Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science. 1999;285:2129–2133. - PubMed
-
- Arron JR, Winslow MM, Polleri A, Chang CP, Wu H, Gao X, Neilson JR, Chen L, Heit JJ, Kim SK, et al. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature. 2006;441:595–600. - PubMed
-
- Baine I, Abe BT, Macian F. Regulation of T-cell tolerance by calcium/NFAT signaling. Immunol Rev. 2009;231:225–240. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

