High-throughput massively parallel sequencing for fetal aneuploidy detection from maternal plasma
- PMID: 23483908
- PMCID: PMC3590217
- DOI: 10.1371/journal.pone.0057381
High-throughput massively parallel sequencing for fetal aneuploidy detection from maternal plasma
Abstract
Background: Circulating cell-free (ccf) fetal DNA comprises 3-20% of all the cell-free DNA present in maternal plasma. Numerous research and clinical studies have described the analysis of ccf DNA using next generation sequencing for the detection of fetal aneuploidies with high sensitivity and specificity. We sought to extend the utility of this approach by assessing semi-automated library preparation, higher sample multiplexing during sequencing, and improved bioinformatic tools to enable a higher throughput, more efficient assay while maintaining or improving clinical performance.
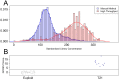
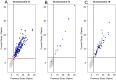
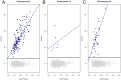
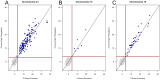
Methods: Whole blood (10mL) was collected from pregnant female donors and plasma separated using centrifugation. Ccf DNA was extracted using column-based methods. Libraries were prepared using an optimized semi-automated library preparation method and sequenced on an Illumina HiSeq2000 sequencer in a 12-plex format. Z-scores were calculated for affected chromosomes using a robust method after normalization and genomic segment filtering. Classification was based upon a standard normal transformed cutoff value of z = 3 for chromosome 21 and z = 3.95 for chromosomes 18 and 13.
Results: Two parallel assay development studies using a total of more than 1900 ccf DNA samples were performed to evaluate the technical feasibility of automating library preparation and increasing the sample multiplexing level. These processes were subsequently combined and a study of 1587 samples was completed to verify the stability of the process-optimized assay. Finally, an unblinded clinical evaluation of 1269 euploid and aneuploid samples utilizing this high-throughput assay coupled to improved bioinformatic procedures was performed. We were able to correctly detect all aneuploid cases with extremely low false positive rates of 0.09%, <0.01%, and 0.08% for trisomies 21, 18, and 13, respectively.
Conclusions: These data suggest that the developed laboratory methods in concert with improved bioinformatic approaches enable higher sample throughput while maintaining high classification accuracy.
Conflict of interest statement
Figures
References
-
- Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, et al. (1997) Presence of fetal DNA in maternal plasma and serum. Lancet 350: 485–487. - PubMed
-
- Nygren AO, Dean J, Jensen TJ, Kruse S, Kwong W, et al. (2010) Quantification of fetal DNA by use of methylation-based DNA discrimination. Clin Chem 56: 1627–1635. - PubMed
-
- Ehrich M, Deciu C, Zwiefelhofer T, Tynan JA, Cagasan L, et al.. (2011) Noninvasive detection of fetal trisomy 21 by sequencing of DNA in maternal blood: a study in a clinical setting. Am J Obstet Gynecol 204: 205 e1–11. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

