(1)H NMR-based metabolite profiling of planktonic and biofilm cells in Acinetobacter baumannii 1656-2
- PMID: 23483923
- PMCID: PMC3590295
- DOI: 10.1371/journal.pone.0057730
(1)H NMR-based metabolite profiling of planktonic and biofilm cells in Acinetobacter baumannii 1656-2
Abstract
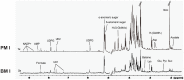
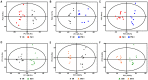
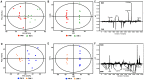
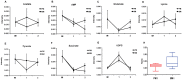
Acinetobacter baumannii is an aerobic and gram-negative pathogenic bacterium that is resistant to most antibiotics. Recently, A. baumannii 1656-2 exhibited the ability to form biofilms under clinical conditions. In this study, global metabolite profiling of both planktonic and biofilm forms of A. baumannii 1656-2 was performed using high-resolution nuclear magnetic resonance (NMR) spectroscopy and multivariate statistical analysis to investigate the metabolic patterns leading to biofilm formation. Principal components analysis (PCA) and orthogonal partial least-squares discriminant analysis (OPLS-DA) score plots showed a distinct separation between planktonic and biofilm cells. Metabolites including acetates, pyruvate, succinate, UDP-glucose, AMP, glutamate, and lysine were increasingly involved in the energy metabolism of biofilm formation. In particular, the ratio of N-acetyl-D-glucosamine (GlcNAc) to D-glucosamine (GlcNH2) was significantly higher during biofilm formation than under the planktonic condition. This study demonstrates that NMR-based global metabolite profiling of bacterial cells can provide valuable insight into the metabolic changes in multidrug resistant and biofilm-forming bacteria such as A. baumannii 1656-2.
Conflict of interest statement
Figures
Similar articles
-
Whole transcriptome analysis of Acinetobacter baumannii assessed by RNA-sequencing reveals different mRNA expression profiles in biofilm compared to planktonic cells.PLoS One. 2013 Aug 30;8(8):e72968. doi: 10.1371/journal.pone.0072968. eCollection 2013. PLoS One. 2013. PMID: 24023660 Free PMC article.
-
Acinetobacter baumannii biofilm biomass mediates tolerance to cold plasma.Lett Appl Microbiol. 2019 Apr;68(4):344-349. doi: 10.1111/lam.13122. Epub 2019 Mar 13. Lett Appl Microbiol. 2019. PMID: 30706947 Free PMC article.
-
[Activity of tigecycline on planktonic and sessile cells of Acinetobacter baumannii].Mikrobiyol Bul. 2009 Oct;43(4):587-95. Mikrobiyol Bul. 2009. PMID: 20084911 Turkish.
-
Analysis of bacterial biofilms using NMR-based metabolomics.Future Med Chem. 2012 Jun;4(10):1273-306. doi: 10.4155/fmc.12.59. Future Med Chem. 2012. PMID: 22800371 Free PMC article. Review.
-
Multivariate analysis of NMR-based metabolomic data.NMR Biomed. 2022 Feb;35(2):e4638. doi: 10.1002/nbm.4638. Epub 2021 Nov 5. NMR Biomed. 2022. PMID: 34738674 Review.
Cited by
-
Anti-Biofilm Activities from Marine Cold Adapted Bacteria Against Staphylococci and Pseudomonas aeruginosa.Front Microbiol. 2015 Dec 14;6:1333. doi: 10.3389/fmicb.2015.01333. eCollection 2015. Front Microbiol. 2015. PMID: 26696962 Free PMC article.
-
Acinetobacter baumannii biofilms: effects of physicochemical factors, virulence, antibiotic resistance determinants, gene regulation, and future antimicrobial treatments.Infect Drug Resist. 2018 Nov 15;11:2277-2299. doi: 10.2147/IDR.S169894. eCollection 2018. Infect Drug Resist. 2018. PMID: 30532562 Free PMC article. Review.
-
Frontiers in superbug management: innovating approaches to combat antimicrobial resistance.Arch Microbiol. 2025 Feb 14;207(3):60. doi: 10.1007/s00203-025-04262-x. Arch Microbiol. 2025. PMID: 39953143 Review.
-
An Overview of Biological and Computational Methods for Designing Mechanism-Informed Anti-biofilm Agents.Front Microbiol. 2021 Apr 13;12:640787. doi: 10.3389/fmicb.2021.640787. eCollection 2021. Front Microbiol. 2021. PMID: 33927701 Free PMC article. Review.
-
Morphological and proteomic analysis of biofilms from the Antarctic archaeon, Halorubrum lacusprofundi.Sci Rep. 2016 Nov 22;6:37454. doi: 10.1038/srep37454. Sci Rep. 2016. PMID: 27874045 Free PMC article.
References
-
- Poirel L, Bonnin RA, Nordmann P (2011) Genetic basis of antibiotic resistance in pathogenic Acinetobacter species. IUBMB Life 63: 1061–1067. - PubMed
-
- Cerqueira GM, Peleg AY (2011) Insights into Acinetobacter baumannii pathogenicity. IUBMB Life 63: 1055–1060. - PubMed
-
- Lee HW, Koh YM, Kim J, Lee J-C, Lee Y-C, et al. (2008) Capacity of multidrug-resistant clinical isolates of Acinetobacter baumannii to form biofilm and adhere to epithelial cell surfaces. Clin Microbiol Infect 14: 49–54. - PubMed
-
- Shin JH, Lee HW, Kim SM, Kim J (2009) Proteomic analysis of Acinetobacter baumannii in biofilm and planktonic growth mode. J Microbiol 47: 728–735. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

