A conformational switch in HP1 releases auto-inhibition to drive heterochromatin assembly
- PMID: 23485968
- PMCID: PMC3907283
- DOI: 10.1038/nature12032
A conformational switch in HP1 releases auto-inhibition to drive heterochromatin assembly
Abstract
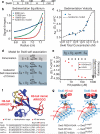
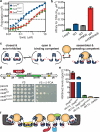
A hallmark of histone H3 lysine 9 (H3K9)-methylated heterochromatin, conserved from the fission yeast Schizosaccharomyces pombe to humans, is its ability to spread to adjacent genomic regions. Central to heterochromatin spread is heterochromatin protein 1 (HP1), which recognizes H3K9-methylated chromatin, oligomerizes and forms a versatile platform that participates in diverse nuclear functions, ranging from gene silencing to chromosome segregation. How HP1 proteins assemble on methylated nucleosomal templates and how the HP1-nucleosome complex achieves functional versatility remain poorly understood. Here we show that binding of the key S. pombe HP1 protein, Swi6, to methylated nucleosomes drives a switch from an auto-inhibited state to a spreading-competent state. In the auto-inhibited state, a histone-mimic sequence in one Swi6 monomer blocks methyl-mark recognition by the chromodomain of another monomer. Auto-inhibition is relieved by recognition of two template features, the H3K9 methyl mark and nucleosomal DNA. Cryo-electron-microscopy-based reconstruction of the Swi6-nucleosome complex provides the overall architecture of the spreading-competent state in which two unbound chromodomain sticky ends appear exposed. Disruption of the switch between the auto-inhibited and spreading-competent states disrupts heterochromatin assembly and gene silencing in vivo. These findings are reminiscent of other conditionally activated polymerization processes, such as actin nucleation, and open up a new class of regulatory mechanisms that operate on chromatin in vivo.
Figures

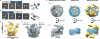
References
-
- Eissenberg JC, Elgin SC. The HP1 protein family: getting a grip on chromatin. Current opinion in genetics & development. 2000;10:204–10. - PubMed
-
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–20. - PubMed
-
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science (New York, NY) 2001;292:110–113. - PubMed
-
- Noma K, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science (New York, NY) 2001;293:1150–1155. - PubMed
-
- Grewal SIS, Jia S. Heterochromatin revisited. Nature reviews Genetics. 2007;8:35–46. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

