Circadian rhythms regulate amelogenesis
- PMID: 23486183
- PMCID: PMC3650122
- DOI: 10.1016/j.bone.2013.02.011
Circadian rhythms regulate amelogenesis
Abstract
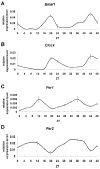
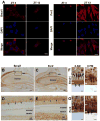
Ameloblasts, the cells responsible for making enamel, modify their morphological features in response to specialized functions necessary for synchronized ameloblast differentiation and enamel formation. Secretory and maturation ameloblasts are characterized by the expression of stage-specific genes which follows strictly controlled repetitive patterns. Circadian rhythms are recognized as key regulators of the development and diseases of many tissues including bone. Our aim was to gain novel insights on the role of clock genes in enamel formation and to explore the potential links between circadian rhythms and amelogenesis. Our data shows definitive evidence that the main clock genes (Bmal1, Clock, Per1 and Per2) oscillate in ameloblasts at regular circadian (24 h) intervals both at RNA and protein levels. This study also reveals that the two markers of ameloblast differentiation i.e. amelogenin (Amelx; a marker of secretory stage ameloblasts) and kallikrein-related peptidase 4 (Klk4, a marker of maturation stage ameloblasts) are downstream targets of clock genes. Both, Amelx and Klk4 show 24h oscillatory expression patterns and their expression levels are up-regulated after Bmal1 over-expression in HAT-7 ameloblast cells. Taken together, these data suggest that both the secretory and the maturation stages of amelogenesis might be under circadian control. Changes in clock gene expression patterns might result in significant alterations of enamel apposition and mineralization.
Copyright © 2013 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures



References
-
- Brown SA, Schibler U. The ins and outs of circadian timekeeping. Curr Opin Genet Dev. 1999;9:588–94. - PubMed
-
- Barclay NL, Eley TC, Mill J, Wong CC, Zavos HM, Archer SN, Gregory AM. Sleep quality and diurnal preference in a sample of young adults: associations with 5HTTLPR, PER3, and CLOCK 3111. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:681–90. - PubMed
-
- Ukai-Tadenuma M, Kasukawa T, Ueda HR. Proof-by-synthesis of the transcriptional logic of mammalian circadian clocks. Nat Cell Biol. 2008;10:1154–63. - PubMed
-
- Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ, Reppert SM. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. - PubMed
-
- Fu L, Patel MS, Bradley A, Wagner EF, Karsenty G. The molecular clock mediates leptin-regulated bone formation. Cell. 2005;122:803–15. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

