RIF1 counteracts BRCA1-mediated end resection during DNA repair
- PMID: 23486525
- PMCID: PMC3630874
- DOI: 10.1074/jbc.M113.457440
RIF1 counteracts BRCA1-mediated end resection during DNA repair
Abstract
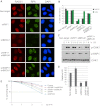
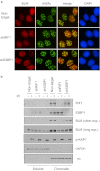
BRCA1 promotes homologous recombination repair and antagonizes 53BP1-dependent nonhomologous end joining (NHEJ) pathway. However, the molecular basis of the competition between BRCA1 and 53BP1 pathways remains elusive. Here we report that RIF1 protein translocates to damage sites via ATM-dependent 53BP1 phosphorylation. Strikingly, loss of RIF1 rescues initial DNA end resection and checkpoint activation in BRCA1-depleted cells. Interestingly RIF1 accumulation at damage sites is antagonized by BRCA1 in S and G2 phases. Conversely, the translocation of BRCA1 to damage sites is inhibited by RIF1 in G1 phase. However, loss of RIF1 differs from that of 53BP1 deficiency, as it cannot fully rescue RAD51 foci formation, homologous recombination defect, and radio-hypersensitivity in BRCA1-deficient cells. This is likely because RIF1, but not 53BP1, also regulates the foci formation and chromatin loading of BLM (the Bloom syndrome helicase). Thus, RIF1 not only acts downstream of 53BP1 and counteracts BRCA1-mediated end resection but also has a secondary role in promoting BLM function in DNA repair.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

