Identification of microRNA targets in tomato fruit development using high-throughput sequencing and degradome analysis
- PMID: 23487304
- PMCID: PMC3638818
- DOI: 10.1093/jxb/ert049
Identification of microRNA targets in tomato fruit development using high-throughput sequencing and degradome analysis
Abstract
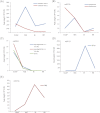
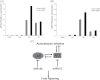
MicroRNAs (miRNAs) play important roles in plant development through regulation of gene expression by mRNA degradation or translational inhibition. Despite the fact that tomato (Solanum lycopersicum) is the model system for studying fleshy fruit development and ripening, only a few experimentally proven miRNA targets are known, and the role of miRNA action in these processes remains largely unknown. Here, by using parallel analysis of RNA ends (PARE) for global identification of miRNA targets and comparing four different stages of tomato fruit development, a total of 119 target genes of miRNAs were identified. Of these, 106 appeared to be new targets. A large part of the identified targets (56) coded for transcription factors. Auxin response factors, as well as two known ripening regulators, colorless non-ripening (CNR) and APETALA2a (SlAP2a), with developmentally regulated degradation patterns were identified. The levels of the intact messenger of both CNR and AP2a are actively modulated during ripening, by miR156/157 and miR172, respectively. Additionally, two TAS3-mRNA loci were identified as targets of miR390. Other targets such as Argonaute 1 (AGO1), shown to be involved in miRNA biogenesis in other plant species, were identified, which suggests a feedback loop regulation of this process. In this study, it is shown that miRNA-guided cleavage of mRNAs is likely to play an important role in tomato fruit development and ripening.
Figures

Similar articles
-
MicroRNA profiling analysis throughout tomato fruit development and ripening reveals potential regulatory role of RIN on microRNAs accumulation.Plant Biotechnol J. 2015 Apr;13(3):370-82. doi: 10.1111/pbi.12297. Epub 2014 Dec 16. Plant Biotechnol J. 2015. PMID: 25516062
-
Ectopic expression of miRNA172 in tomato (Solanum lycopersicum) reveals novel function in fruit development through regulation of an AP2 transcription factor.BMC Plant Biol. 2020 Jun 19;20(1):283. doi: 10.1186/s12870-020-02489-y. BMC Plant Biol. 2020. PMID: 32560687 Free PMC article.
-
Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening.Plant Cell. 2011 Mar;23(3):923-41. doi: 10.1105/tpc.110.081273. Epub 2011 Mar 11. Plant Cell. 2011. PMID: 21398570 Free PMC article.
-
Transcriptional control of fleshy fruit development and ripening.J Exp Bot. 2014 Aug;65(16):4527-41. doi: 10.1093/jxb/eru316. J Exp Bot. 2014. PMID: 25080453 Review.
-
Noncoding RNAs: functional regulatory factors in tomato fruit ripening.Theor Appl Genet. 2020 May;133(5):1753-1762. doi: 10.1007/s00122-020-03582-4. Epub 2020 Mar 24. Theor Appl Genet. 2020. PMID: 32211918 Review.
Cited by
-
Identification of miRNAs and Their Targets Involved in Flower and Fruit Development across Domesticated and Wild Capsicum Species.Int J Mol Sci. 2021 May 4;22(9):4866. doi: 10.3390/ijms22094866. Int J Mol Sci. 2021. PMID: 34064462 Free PMC article.
-
Tomato MicroRNAs and Their Functions.Int J Mol Sci. 2022 Oct 9;23(19):11979. doi: 10.3390/ijms231911979. Int J Mol Sci. 2022. PMID: 36233279 Free PMC article. Review.
-
Tuning LeSPL-CNR expression by SlymiR157 affects tomato fruit ripening.Sci Rep. 2015 Jan 19;5:7852. doi: 10.1038/srep07852. Sci Rep. 2015. PMID: 25597857 Free PMC article.
-
Tomato HAIRY MERISTEM genes are involved in meristem maintenance and compound leaf morphogenesis.J Exp Bot. 2016 Nov;67(21):6187-6200. doi: 10.1093/jxb/erw388. J Exp Bot. 2016. PMID: 27811085 Free PMC article.
-
Comparative Transcriptome Analysis Uncovers the Regulatory Roles of MicroRNAs Involved in Petal Color Change of Pink-Flowered Strawberry.Front Plant Sci. 2022 Mar 29;13:854508. doi: 10.3389/fpls.2022.854508. eCollection 2022. Front Plant Sci. 2022. PMID: 35422831 Free PMC article.
References
-
- Allen E, Xie Z, Gustafson AM, Carrington JC. 2005. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

